Search Count: 65
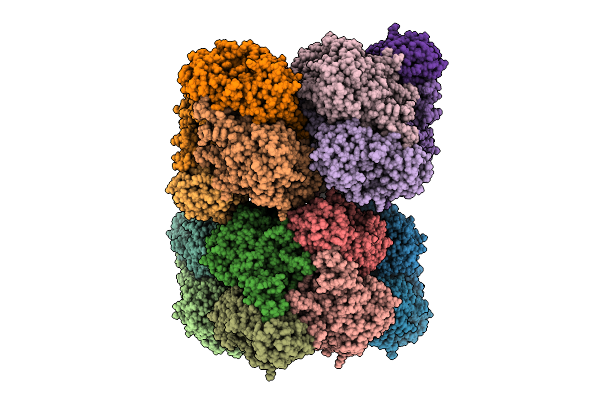 |
Crystal Structure Of Methionine Gamma-Lyase (K209Q Variant) From Brevibacterium Aurantiacum In Complex With Plp And Alpha-Ketobutyrate
Organism: Brevibacterium aurantiacum
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LYASE Ligands: PEG, SO4, 4LM |
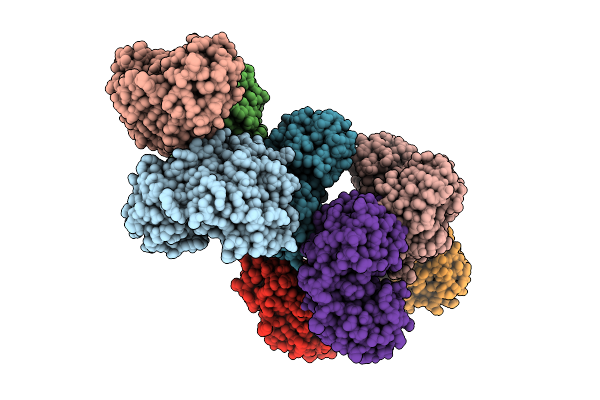 |
Crystal Structure Of Methionine Gamma-Lyase From Brevibacterium Aurantiacum Having Disordered N-Terminus And Devoid Of Plp Cofactor
Organism: Brevibacterium aurantiacum
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LYASE |
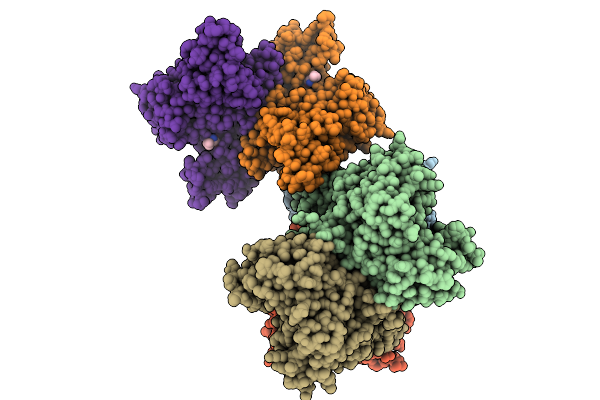 |
Crystal Structure Of Methionine Gamma-Lyase From Brevibacterium Sandarakinum In Complex With Plp And Norleucine At Ph 6.5
Organism: Brevibacterium sandarakinum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: NLE |
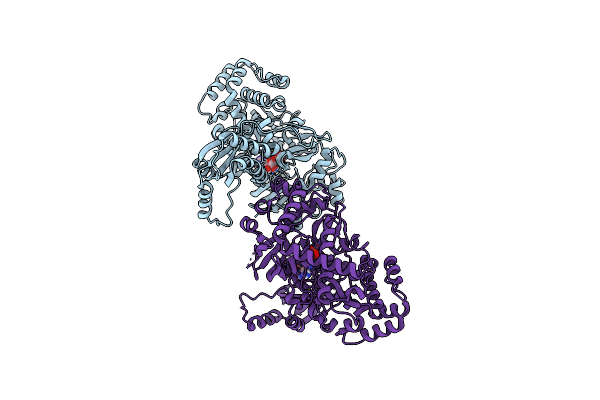 |
Structure Of Indole-3-Acetic Acid-Amido Synthetase Gh3.6 From A.Thaliana In Complex With Amp
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-04-09 Classification: LIGASE Ligands: AMP |
 |
Structure Of Indole-3-Acetic Acid-Amido Synthetase Gh3.6 From A.Thaliana In Complex With Amp And Aspartate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-09 Classification: LIGASE Ligands: AMP, ASP |
 |
Apoform Of Spermine/Spermidine Acetyl Transferase (Atssat) From A. Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-30 Classification: FLAVOPROTEIN Ligands: FAD, EDO, GOL |
 |
Crystal Structure Of Maize Cko/Ckx8 In Complex With Urea-Derived Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-Methoxy-Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: EDO, GOL, FAD, UZ3 |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, UZX |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-(Trifluoromethoxy)Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, V2F |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-Methoxy-Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, UZ3, GOL, EDO |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[[3,5-Dichloro-2-(2-Hydroxyethyl)Phenyl]Carbamoylamino]-4-(Trifluoromethoxy)Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: V1X, FAD |
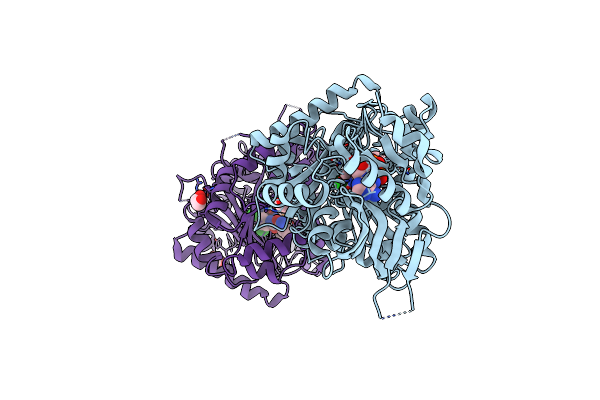 |
Crystal Structure Of Maize Cko/Ckx8 In Complex With Urea-Derived Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-02-21 Classification: FLAVOPROTEIN Ligands: FAD, UZX, GOL, EDO |
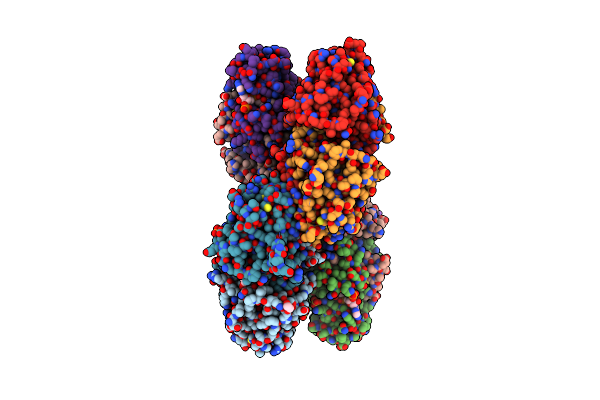 |
Moss Spermine/Spermidine Acetyl Transferase (Ppssat) In Complex With Coa And Lysine
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: COA, LYS, NA, EDO, SO4 |
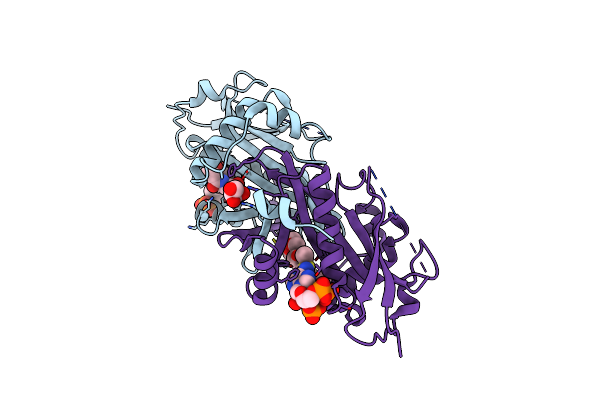 |
Moss Spermine/Spermidine Acetyl Transferase (Ppssat) In Complex With Acetylcoa And Polyethylen Glycol
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-03-15 Classification: TRANSFERASE Ligands: ACO, GOL, 1PE |
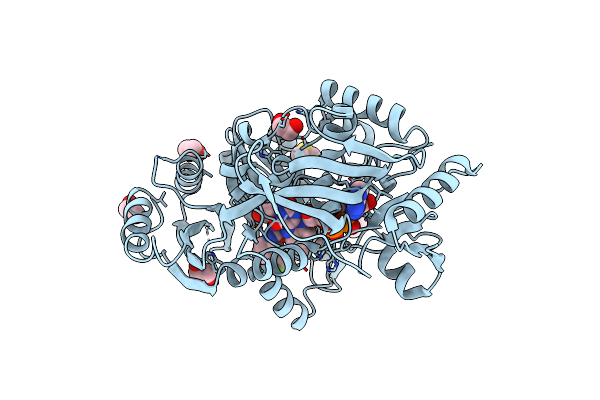 |
Crystal Structure Of Zmcko4A In Complex With Inhibitor 1-[2-(2-Hydroxy-Ethyl)-Phenyl]-3-(3-Trifluoromethoxy-Phenyl)-Urea
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: FLAVOPROTEIN Ligands: FAD, OJ2, EDO, DMS |
 |
Crystal Structure Of Zmcko4A In Complex With Inhibitor 1-(3-Chloro-5-Trifluoromethoxy-Phenyl)-3-[2-(2-Hydroxy-Ethyl)-Phenyl]-Urea
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-10-14 Classification: FLAVOPROTEIN Ligands: EDO, FAD, OHZ, DMS |
 |
Crystal Sttructure Of Zmcko8 In Complex With Inhibitor 1-(3-Chloro-5-Trifluoromethoxy-Phenyl)-3-[2-(2-Hydroxy-Ethyl)-Phenyl]-Urea
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-10-14 Classification: FLAVOPROTEIN Ligands: OHZ, FAD, IPA, PEG, EDO |
 |
Crystal Structure Of Aldehyde Dehydrogenase 21 (Aldh21) From Physcomitrella Patens In Its Apoform
Organism: Physcomitrella patens subsp. patens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: GOL, EDO, PEG |
 |
Crystal Structure Of Aldehyde Dehydrogenase 21 (Aldh21) From Physcomitrella Patens In Complex With The Reaction Product Succinate
Organism: Physcomitrella patens subsp. patens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: SIN, EDO, PEG, GOL |

