Search Count: 24
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-04 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, MG, ACY, GOL, PO4 |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, PO4, MG, ACY, GOL, PEG, NA |
 |
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: MG |
 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease In The Ligand-Free State
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease Bound To Inhibitor Nv-004
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN/INHIBITOR Ligands: GOL, ACT, FHR |
 |
Nmr Shows Why A Small Chemical Change Almost Abolishes The Antimicrobial Activity Of Gccf
Organism: Lactiplantibacillus plantarum
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.83 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: TFA |
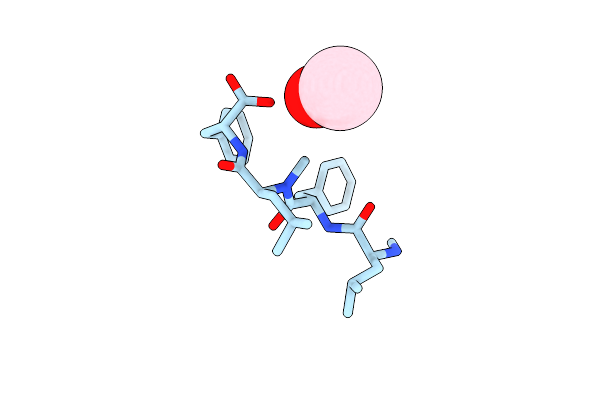 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: MOH |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: HOH |
 |
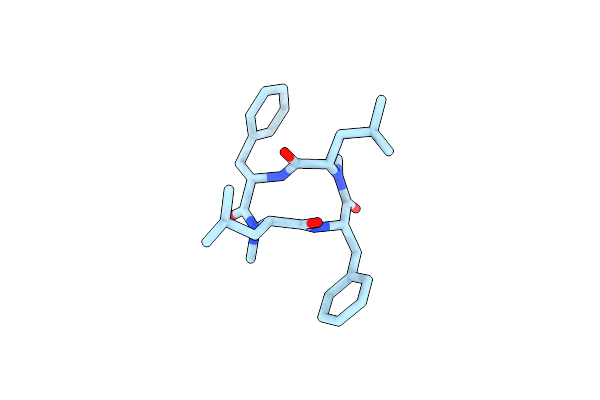
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC |
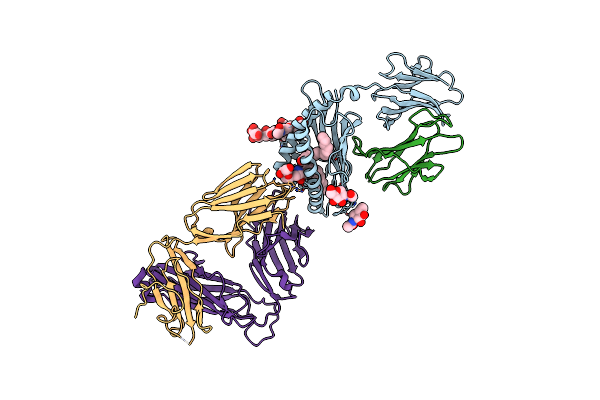 |
Structure Of Alpha-Galactosylphytosphingosine Bound By Cd1D And In Complex With The Va14Vb8.2 Tcr
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2018-04-04 Classification: IMMUNE SYSTEM Ligands: N57, PLM, NAG |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN |
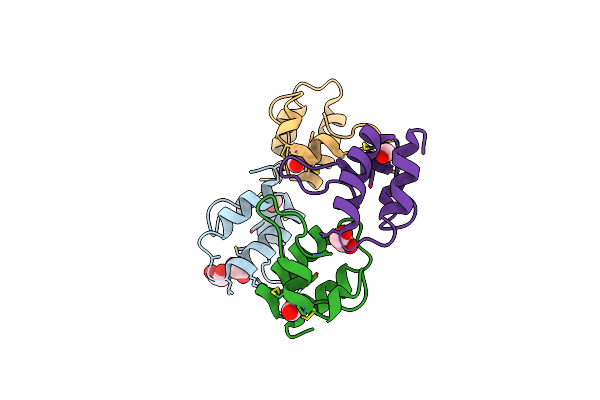 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN Ligands: FMT, EDO |
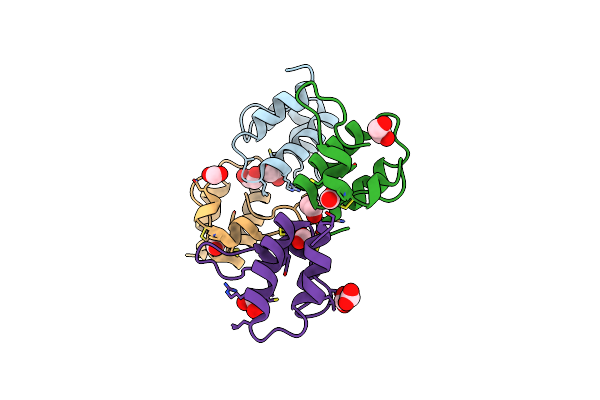 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN Ligands: EDO, FMT |
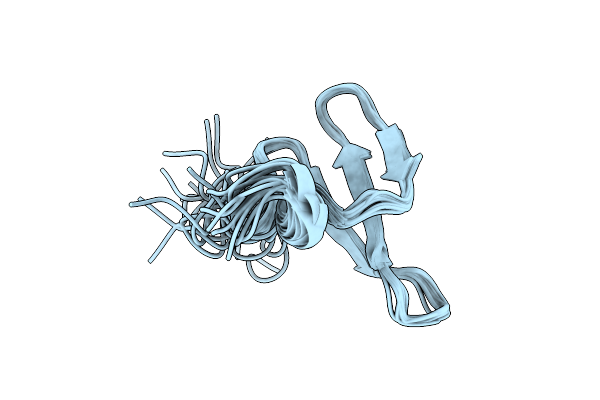 |
 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-09-23 Classification: HYDROLASE |
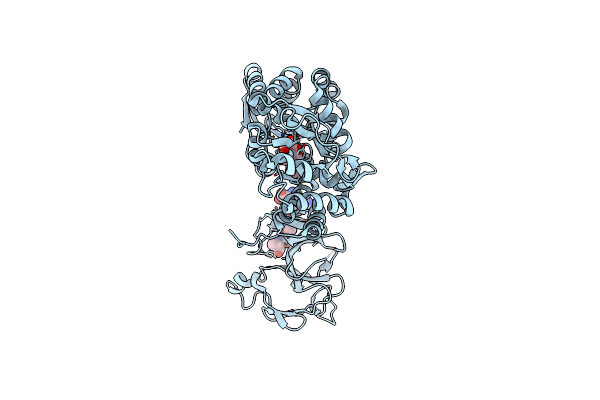 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With A Substrate Peptide (Pep1).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-09-23 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With A Substrate Peptide (Pep2).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-09-23 Classification: HYDROLASE |
 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With An Inhibitor Peptide (Pep3).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-09-23 Classification: hydrolase/hydrolase inhibitor |

