Search Count: 15
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, PEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS, PEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, PEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS, PEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, XSP, TRS, 0V5 |
 |
Fluoride Inhibition Of Enolase: Crystal Structure Of The Inhibitory Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2006-03-21 Classification: LYASE Ligands: MG, PO4, TRS |
 |
Fluoride Inhibition Of Enolase: Crystal Structure Of The Inhibitory Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2006-03-21 Classification: LYASE Ligands: MG, PO4, F, TRS |
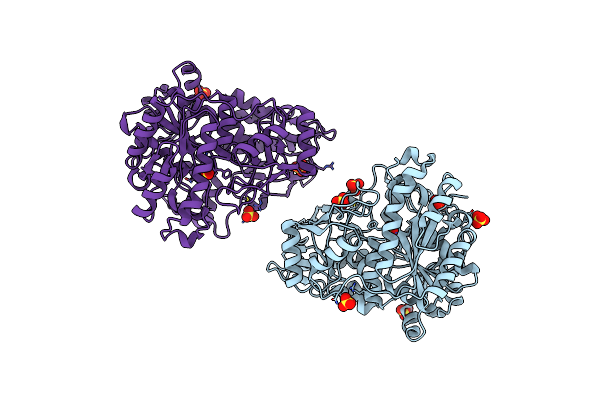 |
Monovalent Cation Binding Sites In N10-Formyltetrahydrofolate Synthetase From Moorella Thermoacetica
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2001-08-31 Classification: LIGASE Ligands: SO4, CS |
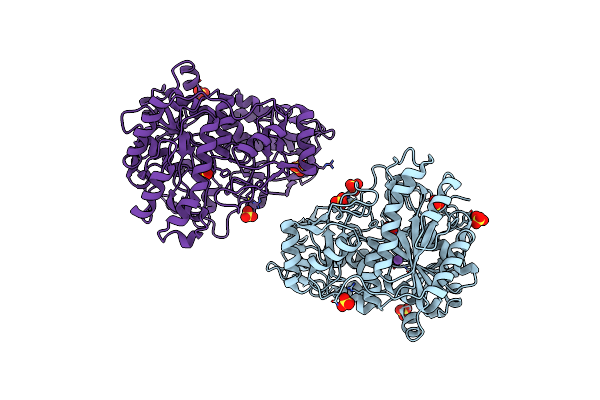 |
Monovalent Cation Binding Sites In N10-Formyltetrahydrofolate Synthetase From Moorella Thermoacetica
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2001-08-30 Classification: LIGASE Ligands: SO4, K |
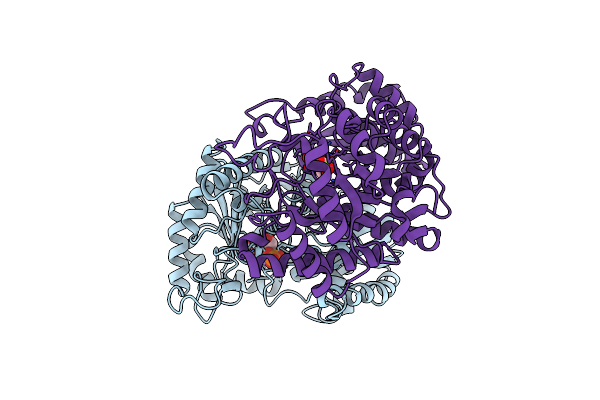 |
Asymmetric Yeast Enolase Dimer Complexed With Resolved 2'-Phosphoglycerate And Phosphoenolpyruvate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-01-14 Classification: LYASE Ligands: MG, LI, 2PG, PEP |
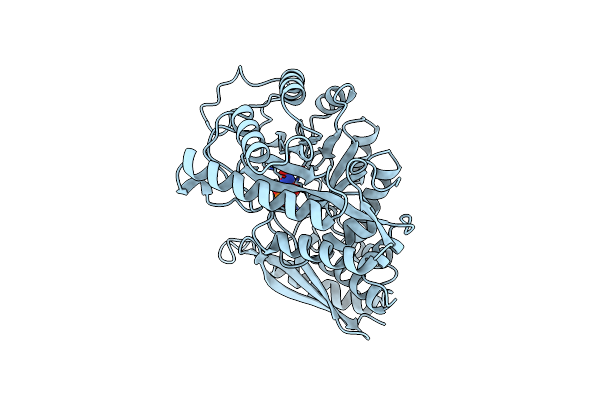 |
Catalytic Metal Ion Binding In Enolase: The Crystal Structure Of Enolase-Mn2+-Phosphonoacetohydroxamate Complex At 2.4 Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-07-31 Classification: CARBON-OXYGEN LYASE Ligands: MN, PAH |
 |
Fluoride Inhibition Of Yeast Enolase: Crystal Structure Of The Enolase-Mg2+-F--Pi Complex At 2.6-Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1994-01-31 Classification: CARBON-OXYGEN LYASE Ligands: MG, PO4, F |
 |
Inhibition Of Enolase: The Crystal Structures Of Enolase-Ca2+-Phosphoglycerate And Enolase-Zn2+-Phosphoglycolate Complexes At 2.2-Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1992-04-15 Classification: CARBON-OXYGEN LYASE Ligands: CA, 2PG |
 |
Inhibition Of Enolase: The Crystal Structures Of Enolase-Ca2+-Phosphoglycerate And Enolase-Zn2+-Phosphoglycolate Complexes At 2.2-Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1992-04-15 Classification: CARBON-OXYGEN LYASE Ligands: PGA, ZN |

