Search Count: 21
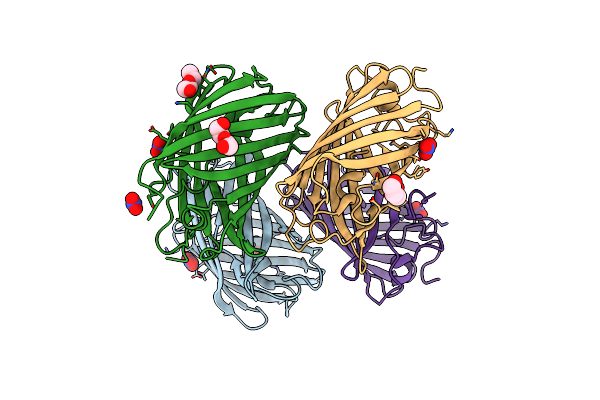 |
Crystal Structure Of A Reconstructed Kaede-Type Red Fluorescent Protein, Lea A69T
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-29 Classification: FLUORESCENT PROTEIN Ligands: NO3, PEG |
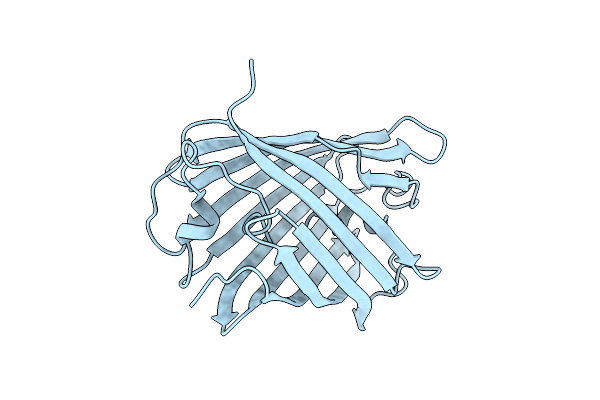 |
Crystal Structure Of A Reconstructed Kaede-Type Red Fluorescent Protein, Lea H62X, Containing 3-Methylhistidine At Position 62
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-29 Classification: LUMINESCENT PROTEIN |
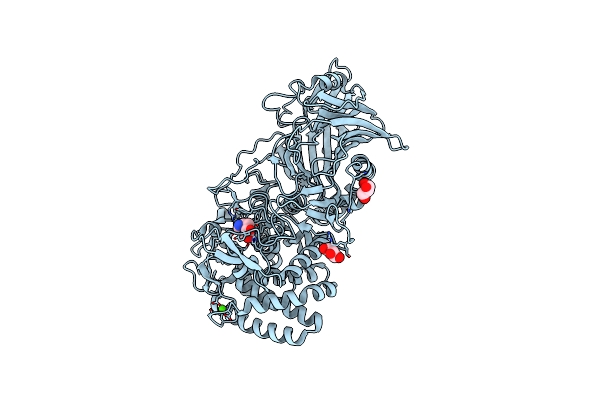 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With A Covalently Bound Cyclophellitol Aziridine
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-22 Classification: HYDROLASE Ligands: PO8, CA, GOL, CL |
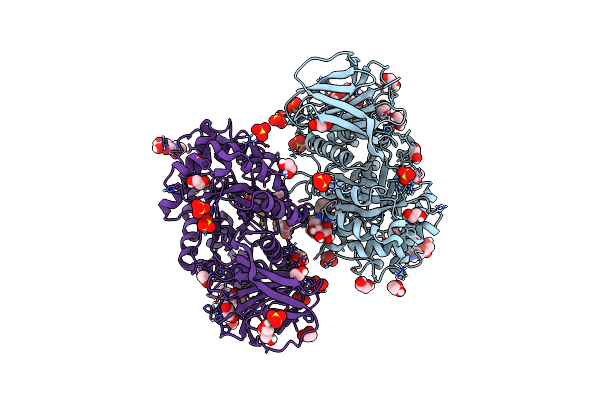 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Bodipy Functionalised Epoxide Activity Based Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: NAG, Q65, K35, SO4, EDO, GOL, GAI, ACT |
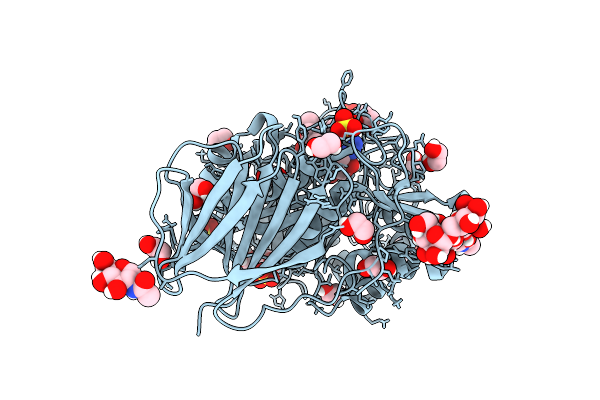 |
Structure Of Recombinant Beta-Glucocerebrosidase In Complex With Bifunctional Cyclophellitol Aziridine Activity Based Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: SO4, NAG, EDO, Q68 |
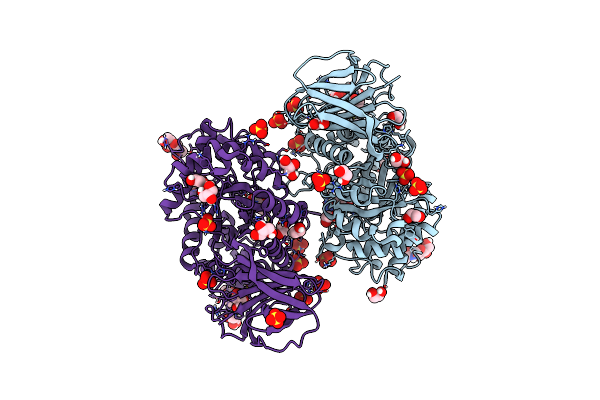 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Azide Tagged Cyclophellitol Epoxide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: NAG, SO4, EDO, GOL, PN8 |
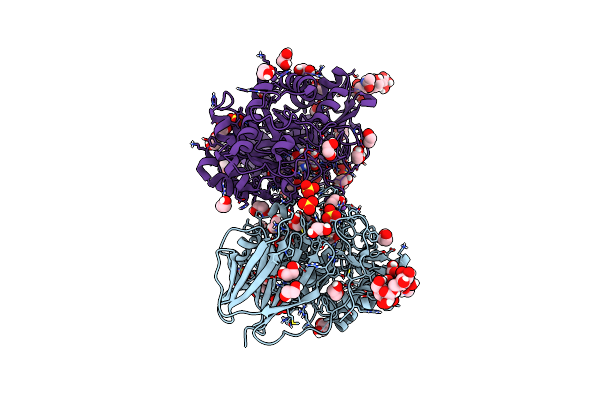 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Cyclophellitol Aziridine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: PO8, SO4, NAG, EDO, NA |
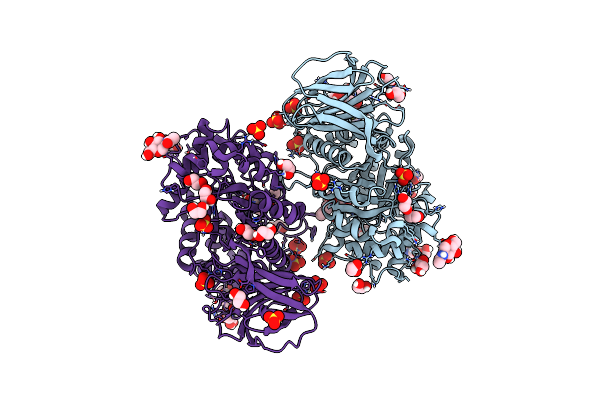 |
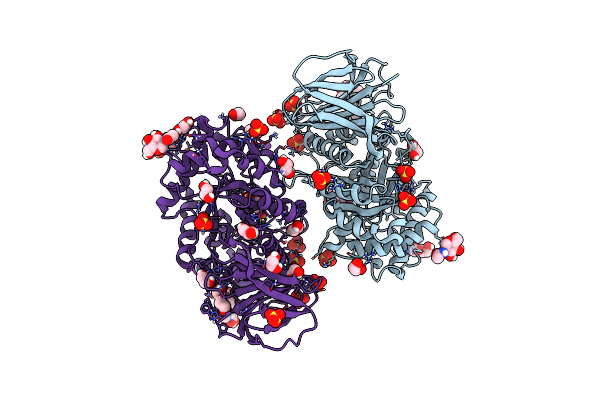
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With N-Acyl Functionalised Cyclophellitol Aziridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: NAG, SO4, EDO, GOL, K35 |
 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Galacto-Configured Cyclophellitol Aziridine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: NAG, SO4, EDO, UUU, GOL, NA |
 |
Crystal Structure Of Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Alpha Cyclophellitol Cyclosulfate Probe Me647
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, PG4, 93Z, OXL |
 |
Crystal Structure Of D412N Nucleophile Mutant Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Unreacted Alpha Cyclophellitol Cyclosulfate Probe Me647
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, EDO, PG4, 94E, OXL |
 |
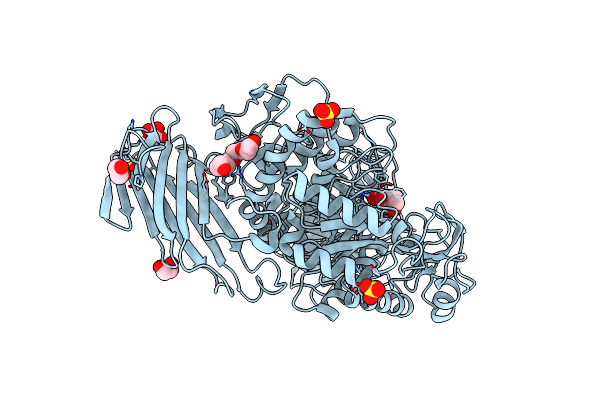
Crystal Structure Of D412N Nucleophile Mutant Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Alpha Cyclophellitol Aziridine Probe Cf021
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, PG4, EDO, 94B, OXL |
 |
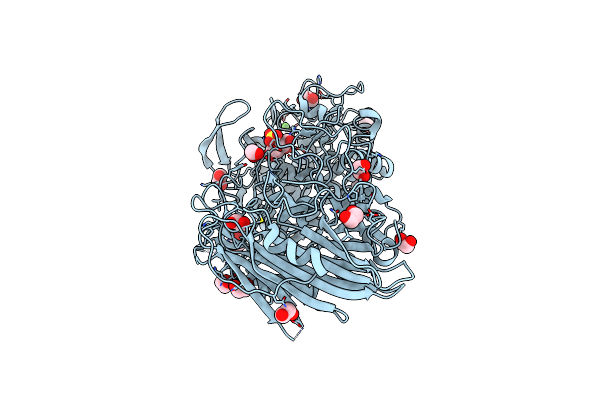
Crystal Structure Of Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Beta Cyclophellitol Aziridine Probe Ky358
Organism: Cellvibrio japonicus (strain ueda107)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, PG4, EDO, 948, OXL |
 |
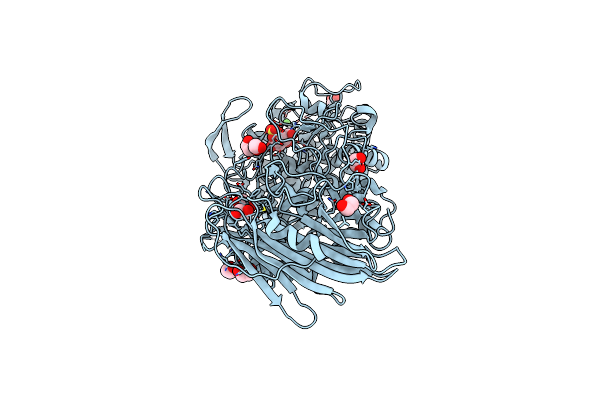
Crystal Structure Of Txgh116 (Beta-Glucosidase From Thermoanaerobacterium Xylolyticum) In Complex With Beta Cyclophellitol Cyclosulfate Probe Me594
Organism: Thermoanaerobacterium xylanolyticum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: EDO, CA, 945 |
 |
Crystal Structure Of Txgh116 (Beta-Glucosidase From Thermoanaerobacterium Xylolyticum) In Complex With Unreacted Beta Cyclophellitol Cyclosulfate Probe Me711
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: EDO, CA, 9FQ |
 |
Crystal Structure Of Thermoanaerobacterium Xylolyticum Gh116 Beta-Glucosidase
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With 2-Deoxy-2-Fluoroglucoside
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: G2F, GOL, EDO, CA |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Deoxynojirimycin
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: NOJ, GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucoimidazole
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GIM, GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: BGC, GOL, EDO, CA |

