Search Count: 249
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: HEM, GOL, A1ITR, DMS, CMO, ACY, SO4, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1I9L, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1JB4, ACT |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Acyl-Coa Dehydrogenase Fade2 Mutant (Pa0508 M130G E296A Q303A) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PGE |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Fade1(Pa0506) E441A From Pseudomonas Aeruginosa Complexed With C16Coa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: PKZ, EDO, PGE, NO3, TRS |
 |
Organism: Escherichia coli (strain k12), Rna interference vector pbsk-gus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: MG |
 |
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-05 Classification: HYDROLASE |
 |
Crystal Structure Of The Apo Acyl-Coa Dehydrogenase Fade2 (Pa0508) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, 1PE, GOL |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Pa0506 (Fade1) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CO8 |
 |
Structural And Functional Analysis Of The Pseudomonas Aeruginosa Pa1677 Protein
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-12-20 Classification: UNKNOWN FUNCTION Ligands: MPD, ACT, LYS |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-10-04 Classification: MEMBRANE PROTEIN Ligands: LFI |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN Ligands: LFI |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN Ligands: ZN, LFI |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN Ligands: ZN, NA, LFI |
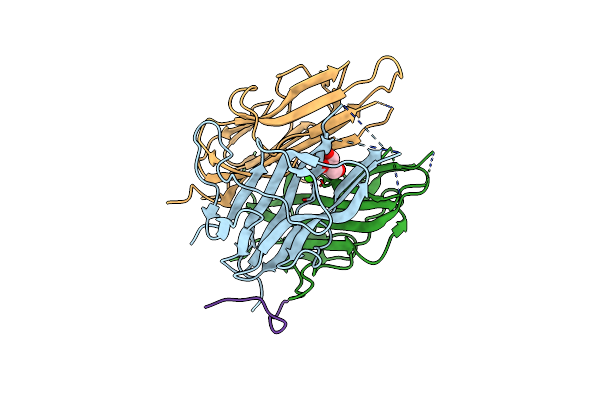 |
High Resolution Crystal Structure Of Exsfa, A Bacillus Cereus Spore Exosporium Protein
Organism: Bacillus cereus, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2023-08-23 Classification: STRUCTURAL PROTEIN Ligands: GOL, CA |
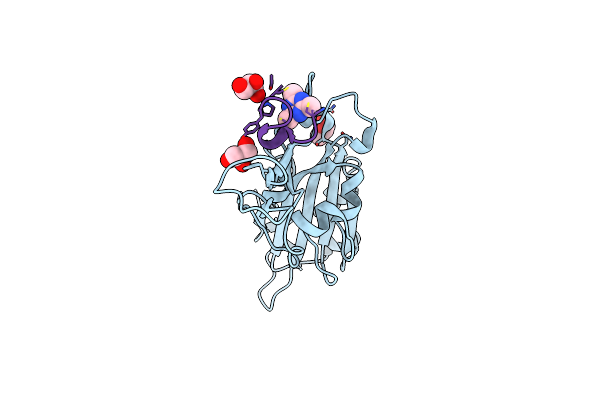 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: GOL, KZ0 |
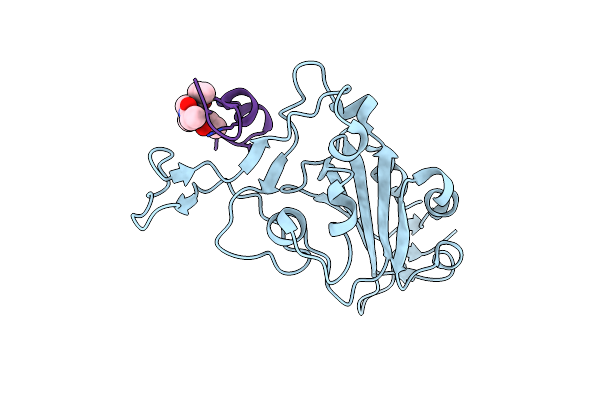 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: 29N |
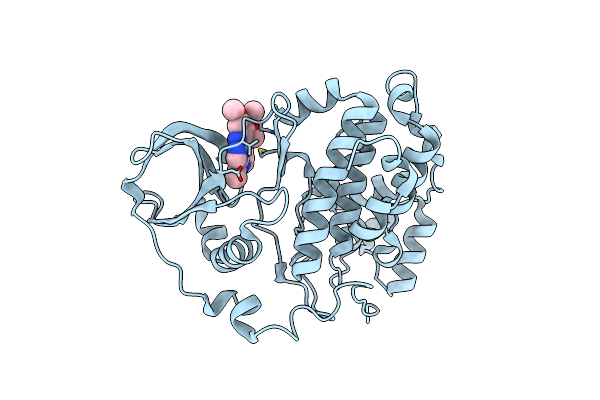 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: QXZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: R7W, EPE, ACT, NA |

