Search Count: 18
 |
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-01-22 Classification: DNA BINDING PROTEIN Ligands: IOD, NO3 |
 |
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-01-22 Classification: DNA BINDING PROTEIN/transcription Ligands: SO4 |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Complexed With Tyrosine And Manganese
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-05-06 Classification: LYASE Ligands: MN, DTY |
 |
Crystal Structure Of The Mutant G226S Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Complexed With Phenylalanine And Manganese
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-04-30 Classification: LYASE Ligands: MN, PHE |
 |
Crystal Structure Of The Tyrosine Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae In Complex With 2-Phosphoglycolate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2004-04-15 Classification: LYASE Ligands: PGA |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-04-15 Classification: LYASE |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae In Complex With Manganese(Ii)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-04-15 Classification: LYASE Ligands: MN |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Complexed With Phenylalanine And Manganese
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-04-15 Classification: LYASE Ligands: MN, PHE |
 |
Double Complex Of The Tyrosine Sensitive Dahp Synthase From S. Cerevisiae With Co2+, Pep And The E4P Analogoue G3P
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-04-08 Classification: LYASE Ligands: CO, PEP, G3P, GOL |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae In Complex With Phosphoenolpyruvate And Cobalt(Ii)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2004-04-08 Classification: LYASE Ligands: CO, PEP, GOL |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae In Complex With Manganese(Ii)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-04-08 Classification: LYASE Ligands: MN, GOL |
 |
Crystal Structure Of The Tyrosine Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae In Complex With Phosphoenolpyruvate And Manganese(Ii)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-01-23 Classification: LYASE Ligands: MN, PEP |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Complexed With Phosphoenolpyruvate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-01-14 Classification: LYASE Ligands: PEP |
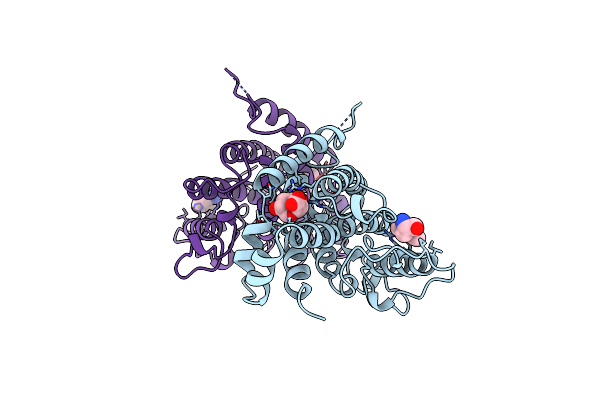 |
Structure Of Yeast Chorismate Mutase With Bound Trp And An Endooxabicyclic Inhibitor
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TRP, TSA |
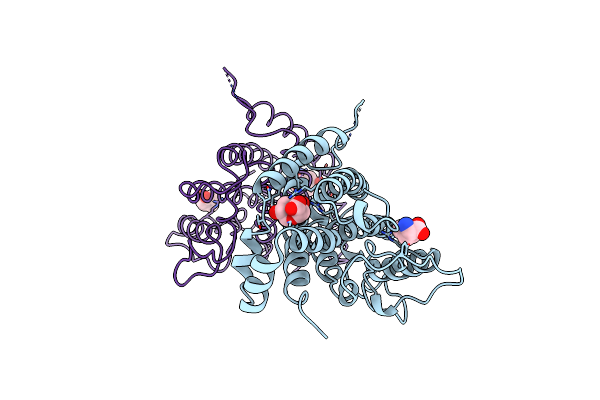 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TYR, TSA |
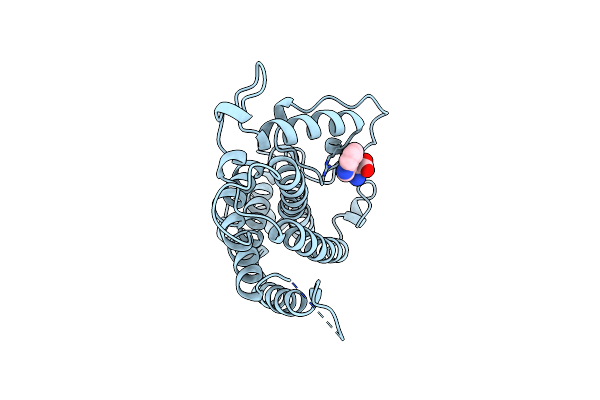 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TRP |
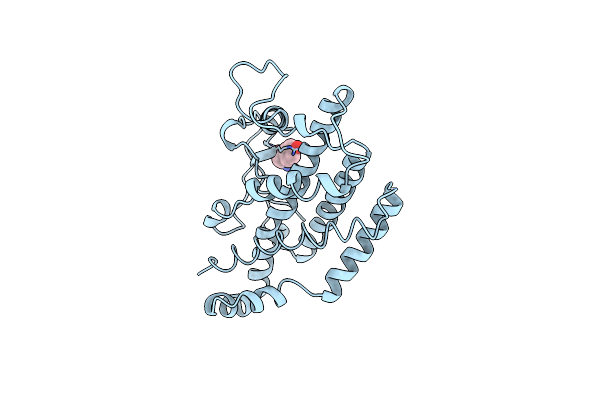 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1996-12-23 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TYR |
 |
The Crystal Structure Of Allosteric Chorismate Mutase At 2.2 Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-09-15 Classification: ISOMERASE Ligands: TRP |

