Search Count: 43
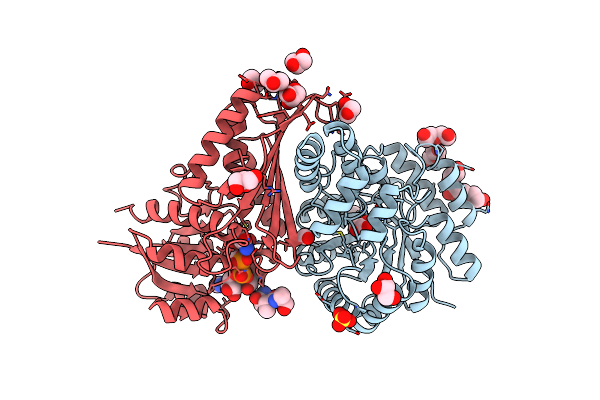 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, PYR, CL, MG, GOL, PEG, NAD, FOR |
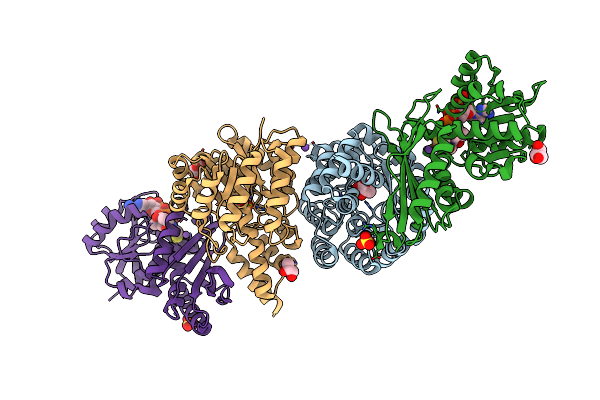 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-09-04 Classification: Lyase/OXIDOREDUCTASE Ligands: PYR, NA, MG, SO4, GOL, COA |
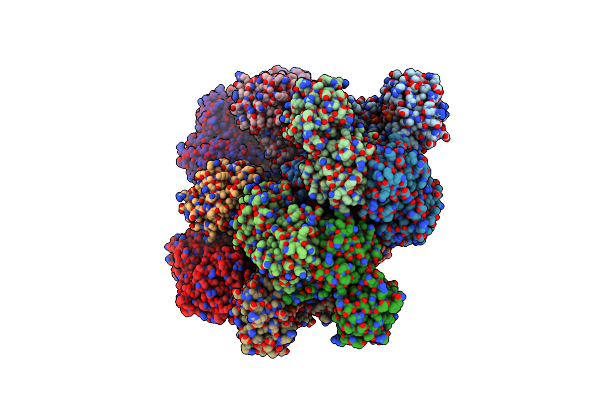 |
Structure Of The D-Erythrose-4-Phosphate Dehydrogenase From E. Coli In Complex With A Nad Cofactor Analog (3-Chloroacetyl Adenine Pyridine Dinucleotide) And Sulfate Anion
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2011-06-08 Classification: OXIDOREDUCTASE Ligands: SO4, 3CD |
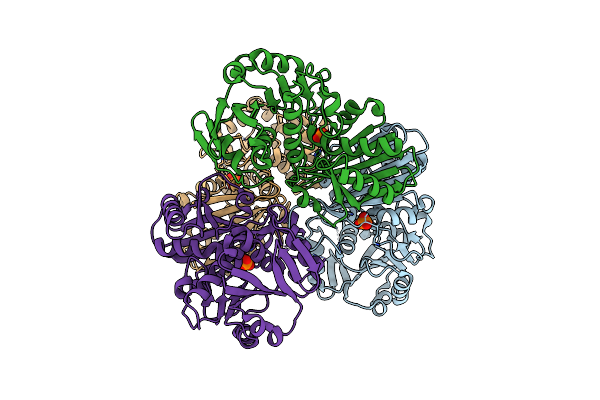 |
Crystal Structure Of The Apoform Of The D-Erythrose-4-Phosphate Dehydrogenase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Structure Of An Active Site Mutant Of The D-Erythrose-4-Phosphate Dehydrogenase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: GOL, EDO, PG4, CL, PGE, PEG |
 |
X-Ray Structure Of Free Methionine-R-Sulfoxide Reductase From Neisseria Meningitidis In Complex With Its Substrate
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2010-05-19 Classification: OXIDOREDUCTASE Ligands: SME, MG, MRD, ACT |
 |
Structure Of The C-Terminal Domain (Msrb) Of Neisseria Meningitidis Pilb (Reduced Form)
Organism: Neisseria meningitidis serogroup a
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Structure Of The C-Terminal Domain (Msrb) Of Neisseria Meningitidis Pilb (Complex With Substrate)
Organism: Neisseria meningitidis serogroup a
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: RSM, CIT, P6G, TRS |
 |
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: ZN, RSM, CA |
 |
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Structural Features Of The Complex Between The Dsbd N-Terminal And The Pilb N-Terminal Domains From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup a, Neisseria meningitidis serogroup b
Method: SOLUTION NMR Release Date: 2009-05-19 Classification: OXIDOREDUCTASE |
 |
Solution Structure Of The C103S Mutant Of The N-Terminal Domain Of Dsbd From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup b
Method: SOLUTION NMR Release Date: 2008-11-11 Classification: OXIDOREDUCTASE |
 |
Solution Structure Of The Oxidized Form (Cys67-Cys70) Of The N-Terminal Domain Of Pilb From N. Meningitidis.
Organism: Neisseria meningitidis serogroup a
Method: SOLUTION NMR Release Date: 2008-07-29 Classification: ELECTRON TRANSPORT |
 |
Solution Structure Of The Reduced Form Of The N-Terminal Domain Of Pilb From N. Meningitidis.
Organism: Neisseria meningitidis serogroup a
Method: SOLUTION NMR Release Date: 2008-07-29 Classification: ELECTRON TRANSPORT |
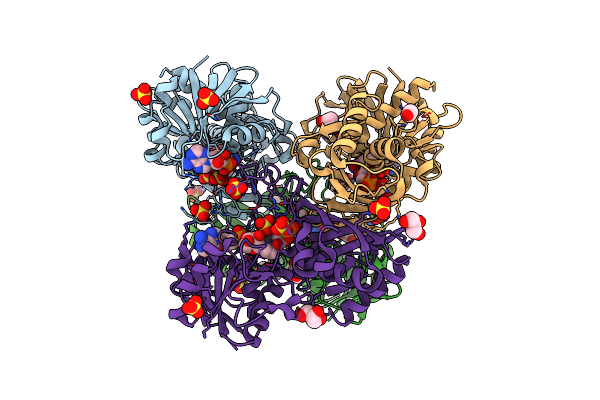 |
Thioacylenzyme Intermediate Of Bacillus Stearothermophilus Phosphorylating Gapdh
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2008-06-17 Classification: OXIDOREDUCTASE Ligands: SO4, G3H, NAD, GOL, EDO |
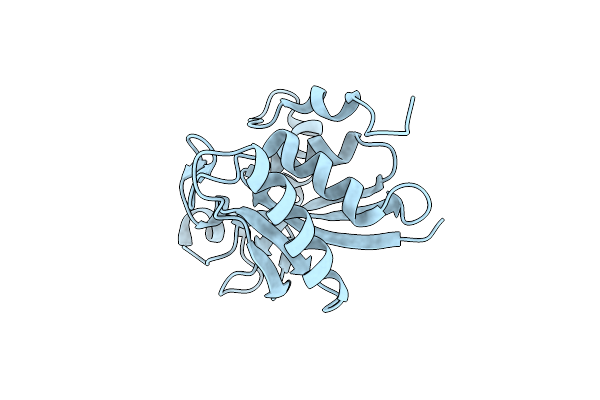 |
Structure Of The Central Domain (Msra) Of Neisseria Meningitidis Pilb (Reduced Form)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-02-19 Classification: OXIDOREDUCTASE |
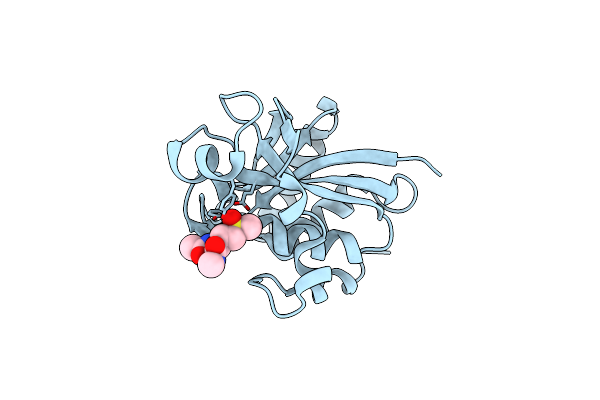 |
Structure Of The Central Domain (Msra) Of Neisseria Meningitidis Pilb (Complex With A Substrate)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2008-02-19 Classification: OXIDOREDUCTASE Ligands: SSM |
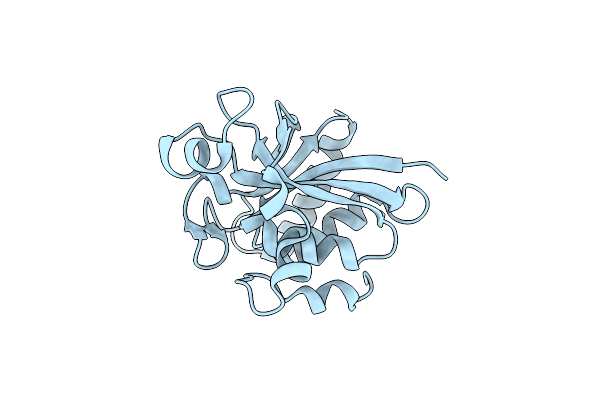 |
Structure Of The Central Domain (Msra) Of Neisseria Meningitidis Pilb (Sulfenic Acid Form)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-02-19 Classification: OXIDOREDUCTASE |
 |
Structure Of The Central Domain (Msra) Of Neisseria Meningitidis Pilb (Oxidized Form)
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-02-19 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Thioacylenzyme Intermediate Of Gapn From S. Mutans, New Data Integration And Refinement.
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2007-12-25 Classification: OXIDOREDUCTASE Ligands: G3H, NAP |

