Search Count: 15
 |
Crystal Structure Of Reverse Transcriptase Domain From Caloramator Australicus Cart-Capp
Organism: Caloramator australicus rc3
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-06-07 Classification: TRANSFERASE Ligands: MG, 1PE, NA, TLA |
 |
Organism: Dysgonamonadaceae bacterium, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-11-30 Classification: DNA BINDING PROTEIN Ligands: BR, GOL |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137
Organism: Marinitoga sp. 1137
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-05-04 Classification: TRANSFERASE |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 With Dgtp
Organism: Marinitoga sp. 1137
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: DGT, MN, EDO |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 With Dsdna
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 - Primer Initiation Complex
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO, DZ4, GTP |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 - Primer Initiation Complex
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO, DZ4, GTP |
 |
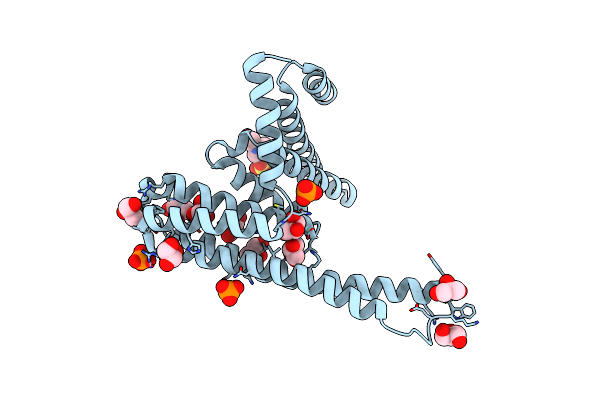
Rigid Body Fitting Of Flagellin Flab, And Flagellar Coiling Proteins, Fcpa And Fcpb, Into A 10 Angstrom Structure Of The Asymmetric Flagellar Filament Purified From Leptospira Biflexa Patoc Wt Cells Resolved Via Subtomogram Averaging
Organism: Leptospira biflexa serovar patoc (strain patoc 1 / atcc 23582 / paris)
Method: ELECTRON MICROSCOPY Release Date: 2020-03-25 Classification: STRUCTURAL PROTEIN |
 |
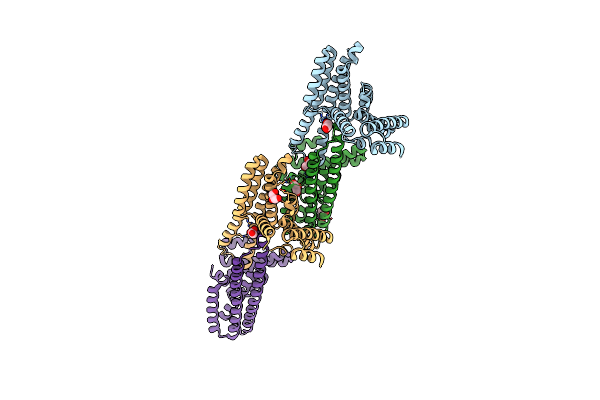
Organism: Leptospira biflexa serovar patoc (strain patoc 1 / atcc 23582 / paris)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-01-29 Classification: PROTEIN FIBRIL Ligands: GOL, PO4, EPE |
 |
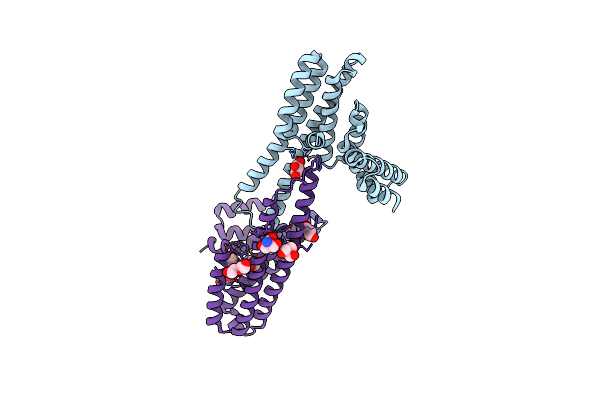
Organism: Leptospira biflexa serovar patoc (strain patoc 1 / atcc 23582 / paris)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-01-29 Classification: PROTEIN FIBRIL Ligands: GOL |
 |
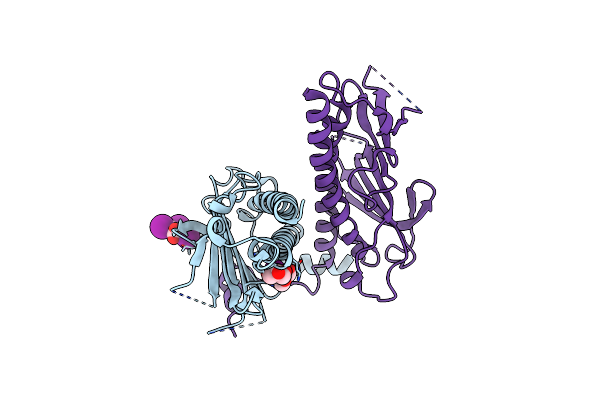
Flagellar Protein Fcpa From Leptospira Biflexa / Ab-Centered Monoclinic Form
Organism: Leptospira biflexa serovar patoc (strain patoc 1 / atcc 23582 / paris)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-29 Classification: PROTEIN FIBRIL Ligands: GOL, IOD, PEG, TRS |
 |
Organism: Leptospira interrogans serogroup icterohaemorrhagiae serovar copenhageni (strain fiocruz l1-130)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2020-01-29 Classification: PROTEIN FIBRIL Ligands: I3C, IOD, GOL |
 |
Nmr Structures Show Unwinding Of The Gcn4P Coiled Coil Superhelix Accompanying Disruption Of Ion Pairs At Acidic Ph
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2017-03-15 Classification: TRANSCRIPTION |
 |
Nmr Structures Show Unwinding Of The Gcn4P Coiled Coil Superhelix Accompanying Disruption Of Ion Pairs At Acidic Ph
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2017-03-15 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2017-03-15 Classification: TRANSCRIPTION |

