Search Count: 15
 |
Crystal Structure Of Reverse Transcriptase Domain From Caloramator Australicus Cart-Capp
Organism: Caloramator australicus rc3
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-06-07 Classification: TRANSFERASE Ligands: MG, 1PE, NA, TLA |
 |
Organism: Dysgonamonadaceae bacterium, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-11-30 Classification: DNA BINDING PROTEIN Ligands: BR, GOL |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137
Organism: Marinitoga sp. 1137
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-05-04 Classification: TRANSFERASE |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 With Dgtp
Organism: Marinitoga sp. 1137
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: DGT, MN, EDO |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 With Dsdna
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 - Primer Initiation Complex
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO, DZ4, GTP |
 |
Prim-Pol Domain Of Crispr-Associated Prim-Pol (Capp) From Marinitoga Sp. 1137 - Primer Initiation Complex
Organism: Marinitoga sp. 1137, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: CO, DZ4, GTP |
 |
Crystal Structure Of Complement C5 In Complex With Chemically Synthesized K92 Knob Domain.
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2022-04-06 Classification: IMMUNE SYSTEM Ligands: EDO |
 |
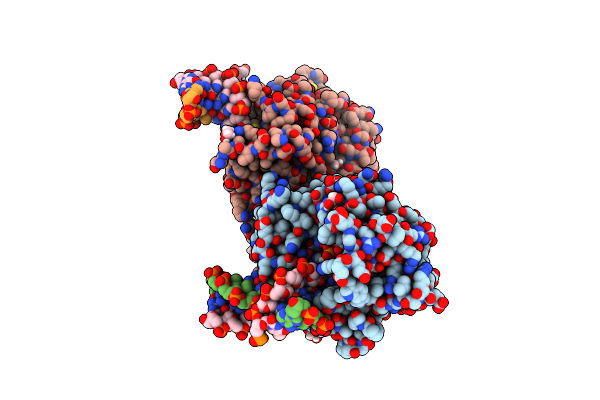
Ternary Complex Of Prim-Polc From Mycobacterium Smegmatis With 2Nt Gapped Dna And Upnhpp
Organism: Mycolicibacterium smegmatis mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: 2KH, MN, CAC, EDO |
 |
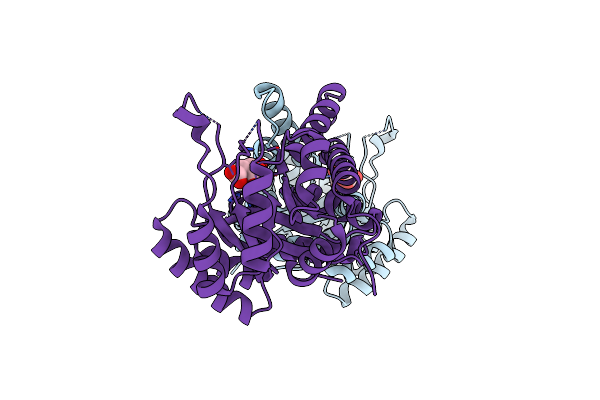
Post Catalytic Complex Of Prim-Polc From Mycobacterium Smegmatis With Gapped Dna And 3'-Dutp
Organism: Mycolicibacterium smegmatis mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: MN, U3H, SO4, MG, EDO, PPV |
 |
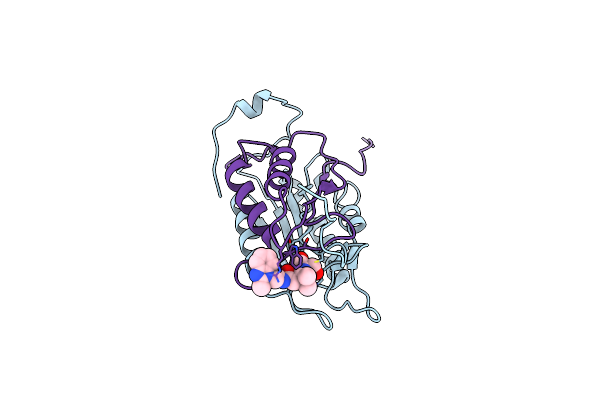
Methicillin-Resistant Staphylococcus Aureus Class Iib Fructose 1,6-Bisphosphate Aldolase
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-11-26 Classification: LYASE Ligands: ZN, FLC |
 |
Succinic Acid Amides As P2-P3 Replacements For Inhibitors Of Interleukin-1Beta Converting Enzyme (Ice Or Caspase 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-08-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3NS |
 |
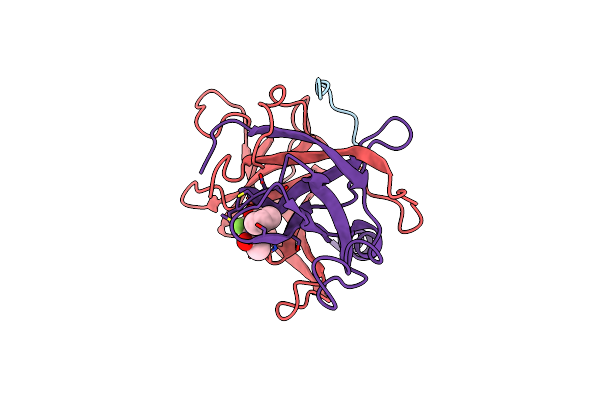
The Structure And Dna Recognition Of A Bifunctional Homing Endonuclease And Group I Intron Splicing Factor
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-01-20 Classification: HYDROLASE/DNA Ligands: MG |
 |
Structure Of Chymotrypsin-*Trifluoromethyl Ketone Inhibitor Complexes. Comparison Of Slowly And Rapidly Equilibrating Inhibitors
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1990-10-15 Classification: HYDROLASE (SERINE PROTEINASE) Ligands: APF |
 |
Structure Of Chymotrypsin-*Trifluoromethyl Ketone Inhibitor Complexes. Comparison Of Slowly And Rapidly Equilibrating Inhibitors
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1990-10-15 Classification: HYDROLASE (SERINE PROTEINASE) Ligands: LPF |

