Search Count: 28
 |
Crystal Structure Of Cep250 Bound To Fkbp12 In The Presence Of Fk506-Like Novel Natural Product
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-29 Classification: CELL CYCLE, ISOMERASE Ligands: 60Z, EDO, PEG, PE8, PGE, PG4, MG, MLA |
 |
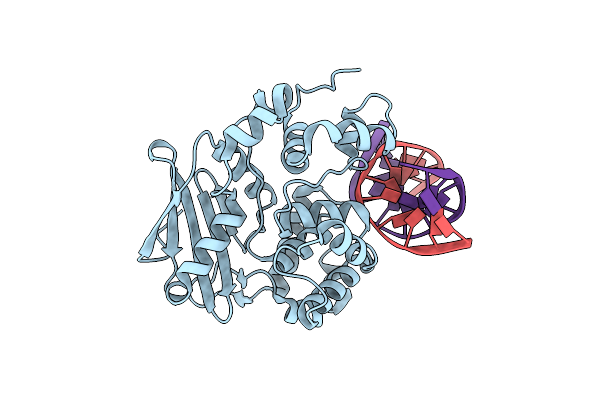
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2012-10-17 Classification: SIGNALING PROTEIN/INHIBITOR |
 |
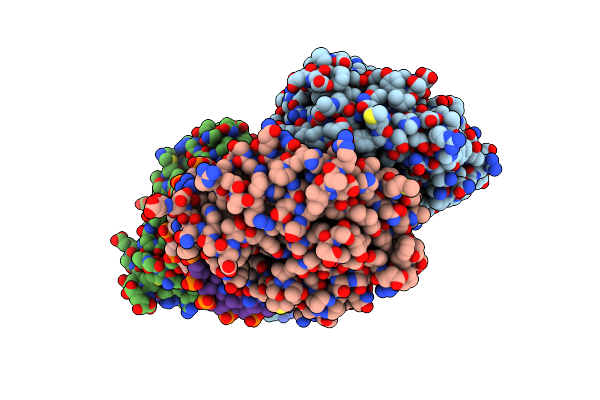
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-09-15 Classification: HYDROLASE/DNA |
 |
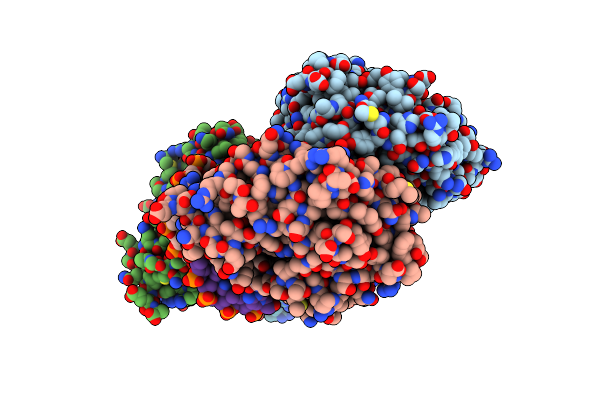
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2010-09-15 Classification: HYDROLASE/DNA |
 |
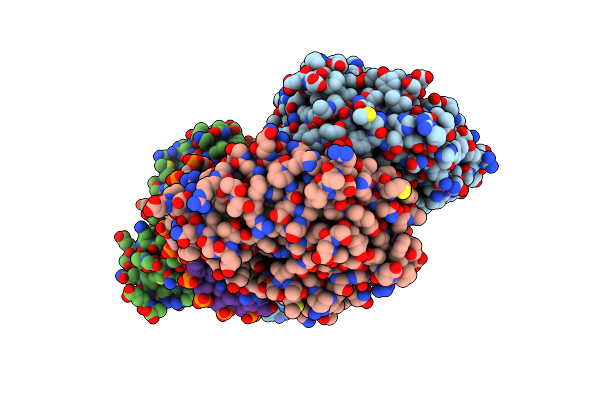
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-09-15 Classification: HYDROLASE/DNA Ligands: HOH |
 |
Crystal Structure Of An Alka Host/Guest Complex N7Methylguanine:Cytosine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-09-09 Classification: HYDROLASE/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 8Oxoguanine:Adenine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 8Oxoguanine:Cytosine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 8Oxoguanine:Cytosine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 8Oxoguanine:Cytosine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 2'-Fluoro-2'-Deoxyinosine:Thymine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 2'-Fluoro-2'-Deoxyinosine:Adenine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of An Alka Host/Guest Complex 2'-Fluoro-2'-Deoxy-1,N6-Ethenoadenine:Thymine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-09-02 Classification: hydrolase/DNA |
 |
Crystal Structure Of Bacillus Stearothermophilus Uvrc 5' Endonuclease Domain
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-08-05 Classification: HYDROLASE |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2008-01-08 Classification: HYDROLASE Ligands: ADP, ZN |
 |
Structure Of The C-Terminal Dimerization Domain Of Infectious Bronchitis Virus Nucleocapsid Protein
Organism: Infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-06-27 Classification: VIRUS/VIRAL PROTEIN/RNA BINDING PROTEIN |
 |
Structure Of The C-Terminal Dimerization Domain Of Infectious Bronchitis Virus Nucleocapsid Protein
Organism: Infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-06-27 Classification: VIRUS/VIRAL PROTEIN/RNA BINDING PROTEIN |
 |
Structure Of The N-Terminal Domain Of Avian Infectious Bronchitis Virus Nucleocapsid Protein (Strain Gray) In A Novel Dimeric Arrangement
Organism: Infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-06-27 Classification: VIRAL PROTEIN |
 |
X-Ray Structure Of The N And C-Terminal Domain Of Coronavirus Nucleocapsid Protein.
Organism: Avian infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2006-06-21 Classification: NUCLEOCAPSID PROTEIN |
 |
Organism: Avian infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-06-19 Classification: VIRAL PROTEIN |

