Search Count: 9
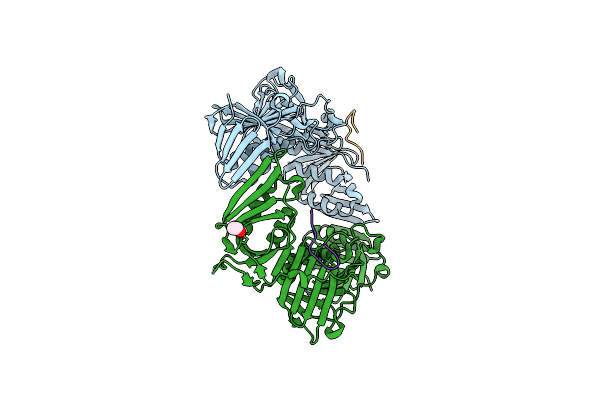 |
Crystal Structure Of Dna Polymerase Iii Subunit Beta From Mycobacterium Marinum In Complex With A Natural Product
Organism: Mycobacterium marinum (strain atcc baa-535 / m), Streptomyces caelicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: EDO |
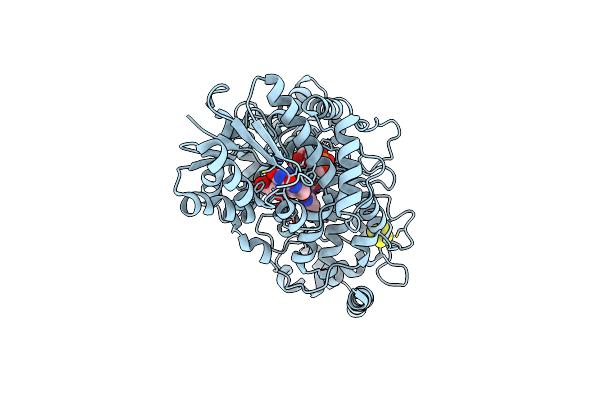 |
Crystal Structure Of Bacterial (6-4) Photolyase Phrb From In Situ Serial Laue Diffraction
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-07-18 Classification: LYASE Ligands: FAD, DLZ, SF4 |
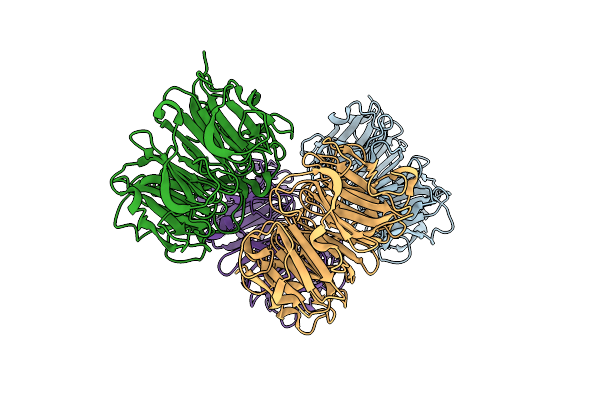 |
Crystal Structure Of Plant Uvb Photoreceptor Uvr8 From In Situ Serial Laue Diffraction
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-18 Classification: GENE REGULATION |
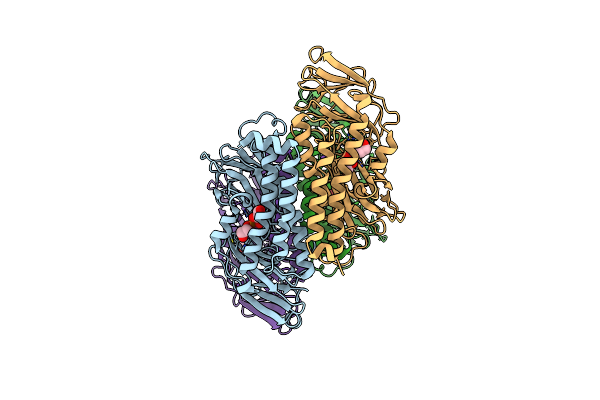 |
Crystal Structure Of A Putative Uncharacterized Protein From Burkholderia Cenocepacia
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-30 Classification: UNKNOWN FUNCTION Ligands: PO4, NA, GOL, EDO |
 |
Organism: Neisseria gonorrhoeae (strain nccp11945)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-30 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of Arginyl-Trna_Synthetase From Neisseria Gonorrhoeae In Complex With Arginine
Organism: Neisseria gonorrhoeae (strain nccp11945)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-08-30 Classification: LIGASE Ligands: ARG, GOL |
 |
Crystal Structure Of Dna Polymerase Iii Subunit Beta From Rickettsia Conorii
Organism: Rickettsia conorii (strain atcc vr-613 / malish 7)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: EDO, MRD |
 |
Crystal Structure Of Iron-Sulfur Cluster Containing Photolyase Phrb Mutant I51W
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: LYASE Ligands: FAD, SF4, GOL |
 |
Crystal Structure Of Iron-Sulfur Cluster Containing Bacterial (6-4) Photolyase Phrb - Y424F Mutant With Impaired Dna Repair Activity
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-01-11 Classification: LYASE Ligands: FAD, DLZ, SF4, GOL |

