Search Count: 151
 |
Aplysia Californica Acetylcholine-Binding Protein In Complex With Spiroimine (+)-4 R
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-03 Classification: CHOLINE-BINDING PROTEIN Ligands: ILR, NAG, CL |
 |
Aplysia Californica Acetylcholine-Binding Protein In Complex With Spiroimine (-)-4 S
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-03 Classification: CHOLINE-BINDING PROTEIN Ligands: WSP, CL, IPA |
 |
Aplysia Californica Acetylcholine-Binding Protein In Complex With Racemic Spiroimine (+)/(-)-4
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-03 Classification: CHOLINE-BINDING PROTEIN Ligands: ILR, IPA |
 |
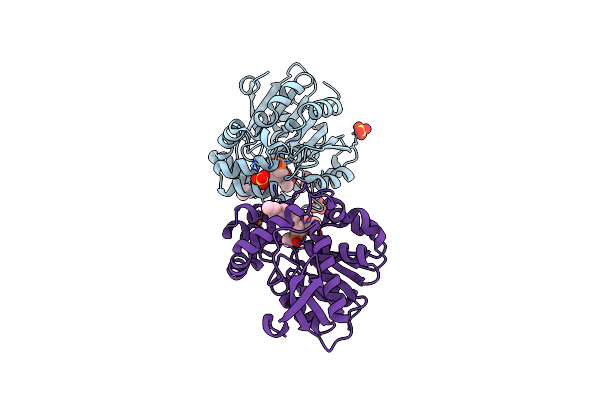
Crystal Structure Of Hsad From Mycobacterium Tuberculosis At 1.96 A Resolution
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: EDO, CL |
 |
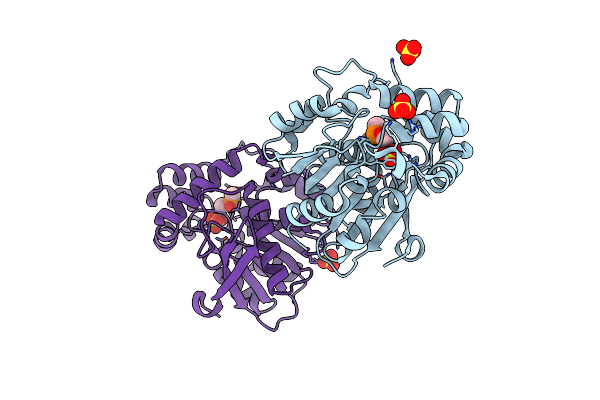
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclophostin-Like Inhibitor Cyc7B
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: IY8, SO4 |
 |
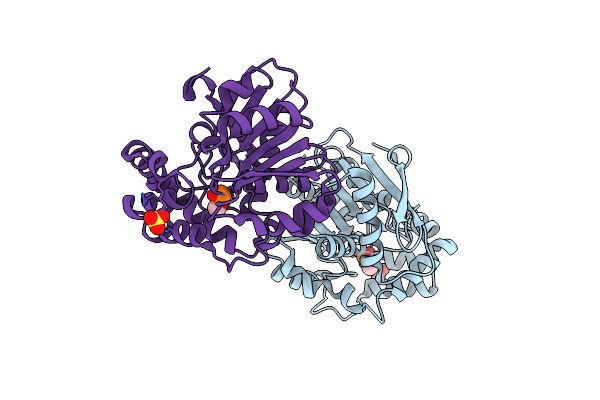
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclophostin-Like Inhibitor Cyc8B
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: 9SW, SO4 |
 |
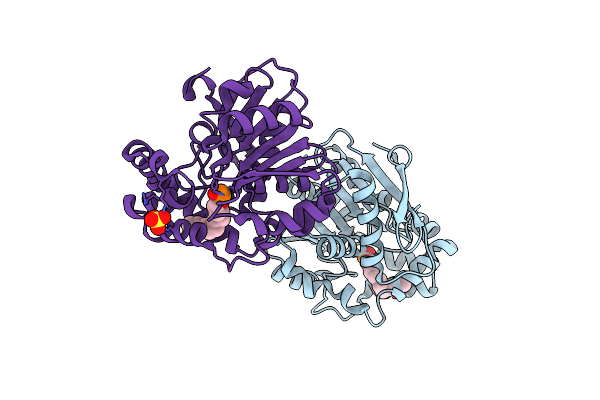
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclipostin-Like Inhibitor Cyc17
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: E9H, SO4 |
 |
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclipostin-Like Inhibitor Cyc31
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: IYB, SO4 |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2022-07-13 Classification: SIGNALING PROTEIN Ligands: CA, PGR |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-07-06 Classification: SIGNALING PROTEIN Ligands: CD, BEF, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-07-06 Classification: SIGNALING PROTEIN Ligands: CD, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: NAG, ACT, SO4, CA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: NAG, GOL, ACT, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: CA, NAG, GOL, MES, P6G |
 |
Structural Insights Into The Periplasmic Sensor Domain Of The Gacs Histidine Kinase Controlling Biofilm Formation In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2018-02-07 Classification: SIGNALING PROTEIN |
 |
Structure Of Maf Glycosyltransferase From Magnetospirillum Magneticum Amb-1
Organism: Magnetospirillum magneticum amb-1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-15 Classification: TRANSFERASE Ligands: SO4, CL, PGE, PG4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: SO4, CL, PGE, PEG, EDO |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With N-Acetyl-Cysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: SC2, SO4, CL, PGE, GOL, EDO |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With 1-Deoxynojirimycin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: NOJ, SO4, CL, EDO |

