Search Count: 24
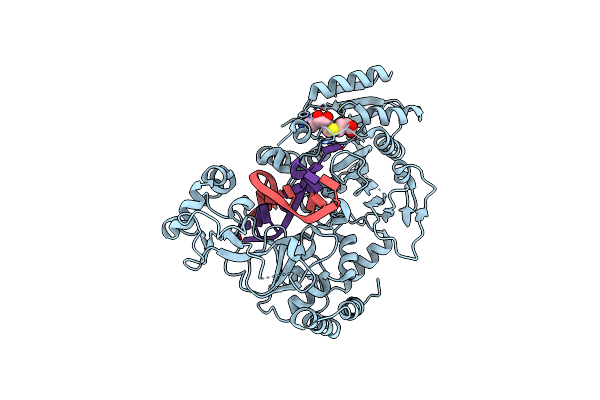 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Unmethylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
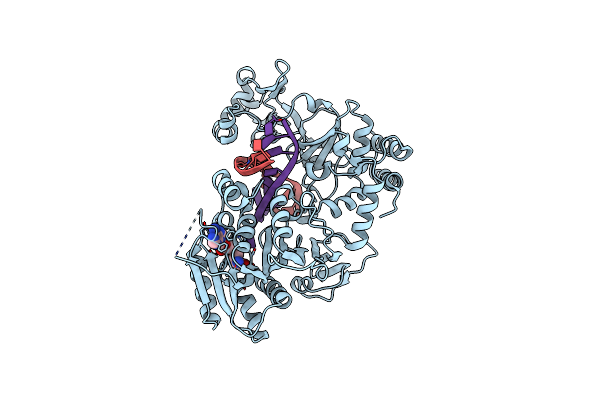 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Hemimethylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Fully Methylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-25 Classification: TRANSFERASE |
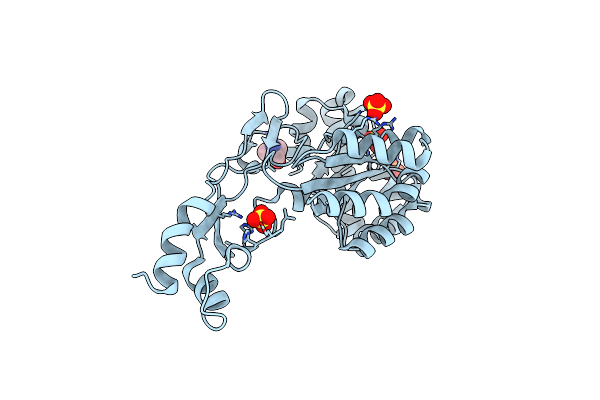 |
Crystal Structure Of Wt Ba3943, A Ce4 Family Pseudoenzyme From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2021-05-19 Classification: UNKNOWN FUNCTION Ligands: SO4, TRS |
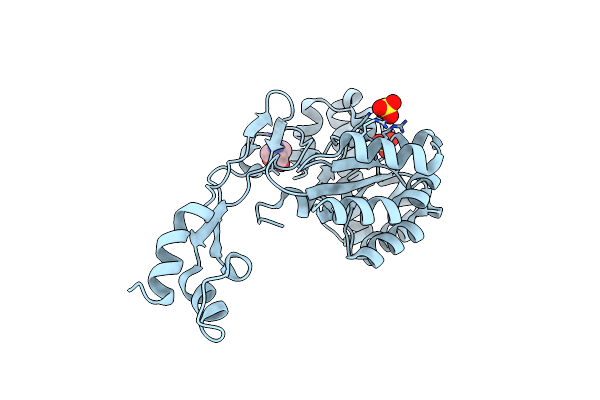 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-10-09 Classification: UNKNOWN FUNCTION Ligands: SO4, TRS |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: ZN, ACT, EDO, SO4 |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, EDO |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With N-Hydroxynaphthalene-1-Carboxamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: 8GK, ZN, EDO, NA, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (E)-N-Hydroxy-3-(Naphthalen-1-Yl)Prop-2-Enamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ACT, ZN, EDO, 8SQ, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (2S)-2,6-Diamino-N-Hydroxyhexanamide
Organism: Bacillus cereus atcc 14579, Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: 8SZ, ZN, CIT, EDO, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (2S)-2-Amino-5-(Diaminomethylideneamino)-N-Hydroxypentanamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ACT, ZN, AHL, CIT |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With Acetazolamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, AZM, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With Thiametg
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, NHT, PGE, EDO, CIT |
 |
Alcohol Dehydrogenase From The Antarctic Psychrophile Moraxella Sp. Tae 123
Organism: Moraxella sp. (strain tae123)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Crystal Structure Of The Putative Polysaccharide Deacetylase Ba0330 From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2015-04-08 Classification: HYDROLASE Ligands: ACT, ZN, EDO |
 |
Crystal Structure Of The Bc1960 Peptidoglycan N-Acetylglucosamine Deacetylase From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: SO4, ACT |
 |
Structural Determination Of Ba0150, A Polysaccharide Deacetylase From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-02-12 Classification: HYDROLASE Ligands: PGE |
 |
Crystal Structure Of Bc0361, A Polysaccharide Deacetylase From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-10 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Structure Of Tab5 Alkaline Phosphatase Mutant His 135 Asp With Mg Bound In The M3 Site.
Organism: Antarctic bacterium tab5
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2009-11-24 Classification: HYDROLASE Ligands: ZN, MG |

