Search Count: 24
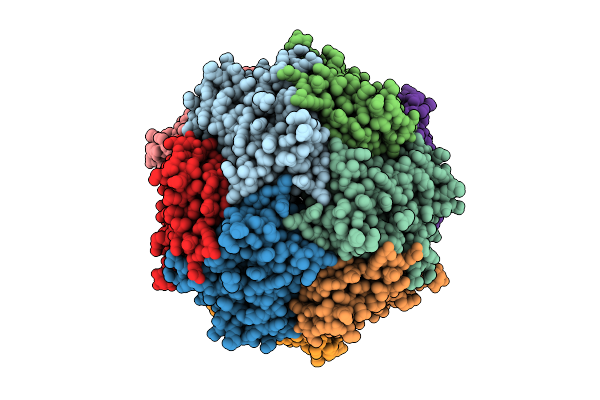 |
Structure Of Thioferritin (Pfdpsl) With Ferrihydrite Growth At A Single Three-Fold Pore.
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: FE, OXY, O |
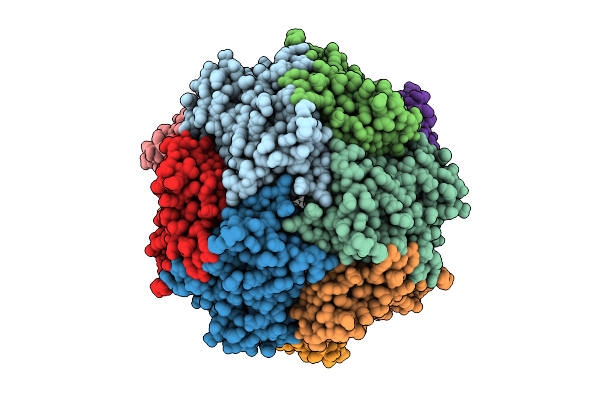 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: METAL BINDING PROTEIN Ligands: FE |
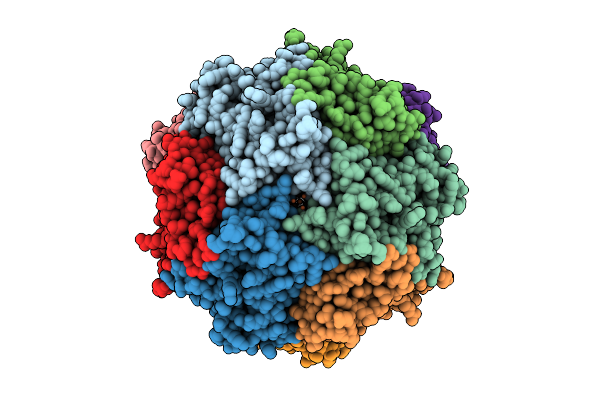 |
Structure Of Thioferritin Exhibiting Iron Mineral Nucleation, From Pyrococcus Furiosis
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: METAL BINDING PROTEIN Ligands: FE, O |
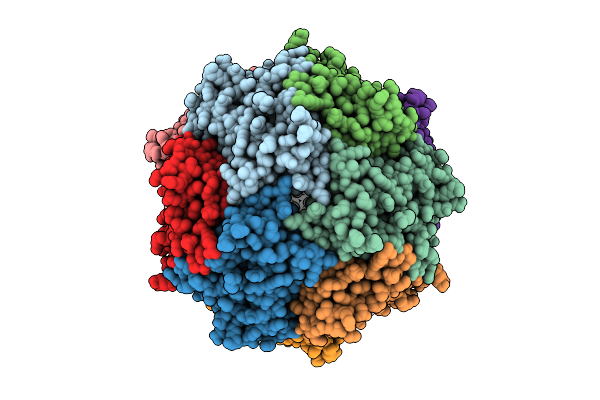 |
Structure Of Thioferritin With Averaged Iron Mineral Core, From Pyrococcus Furiosis
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.68 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Cryoem Structure Of Bchn-Bchb Bound To Pchlide From The Dpor Under Turnover Complex Dataset
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.29 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Cryoem Structure Of Bchn-Bchb Electron Acceptor Component Protein Of Dpor With Pchlide
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, CU |
 |
Cryoem Structure Of Bchn-Bchb Electron Acceptor Component Protein Of Dpor With Pchlide
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.13 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: RECOMBINATION |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: FES, FE |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: FES, SO4, UB7, FE |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: SO4, FES, FE, 8IB |
 |
Dark-Operative Protochlorophyllide Oxidoreductase In The Nucleotide-Free Form.
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: SF4, CL |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-03-14 Classification: SIGNALING PROTEIN Ligands: MPD, NI |
 |
Organism: Pyrococcus furiosus com1
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, MG |
 |
Nadp(H) Bound Nadh-Dependent Ferredoxin:Nadp Oxidoreductase (Nfni) From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, NDP, GOL, FES, MG |
 |
Nadh-Dependent Ferredoxin:Nadp Oxidoreductase (Nfni) From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, MG |
 |
[Fefe]-Hydrogenase Oxygen Inactivation Is Initiated By The Modification And Degradation Of The H Cluster 2Fe Subcluster
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: SF4, CL, ARS |

