Search Count: 51
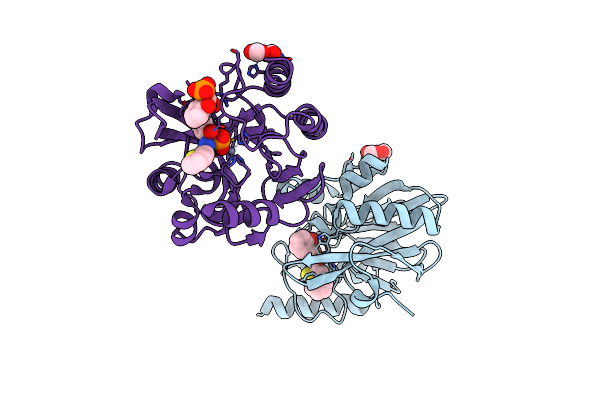 |
Vim-2 In Complex With Gkv61 (5C) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IKA, ZN, A1H8T |
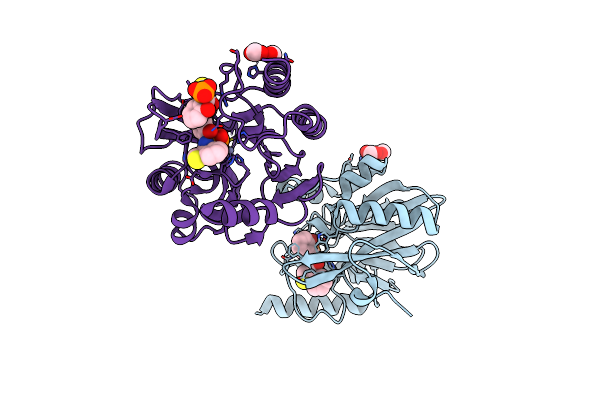 |
Vim-2 In Complex With Gkv53 (5D) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IJ9, ZN, A1H8W |
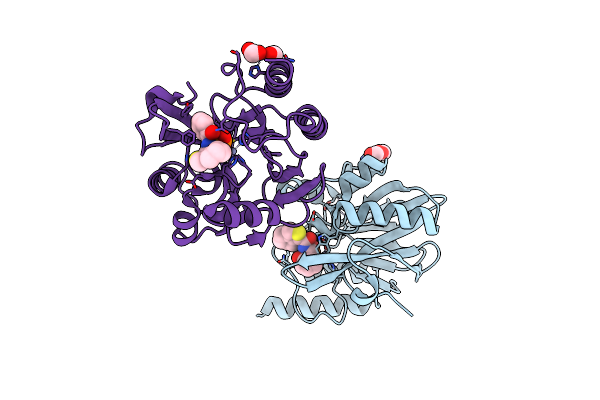 |
Vim-2 In Complex With Gkv65 (5G) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8V, ZN, MG |
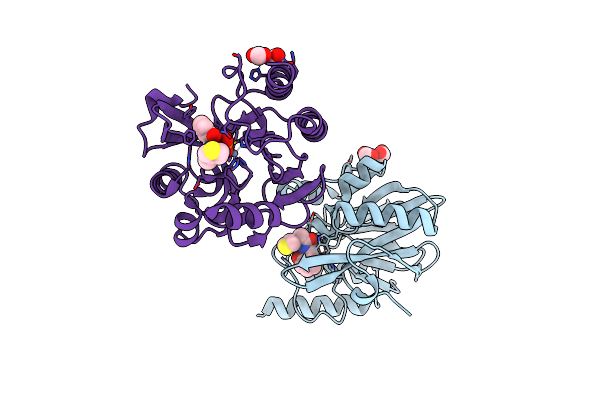 |
Vim-2 In Complex With Gkv63 (5J) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8U, ZN |
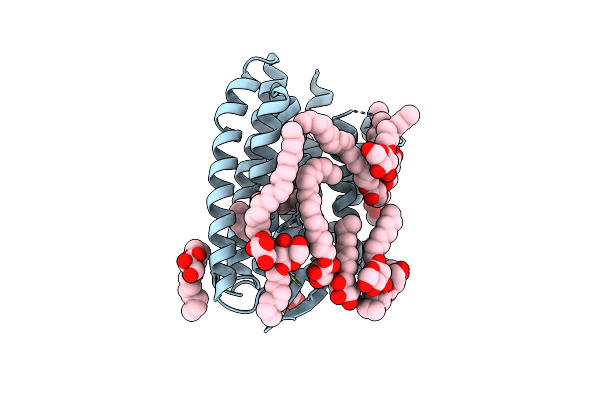 |
Grouped 150-240 Ms Dark Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
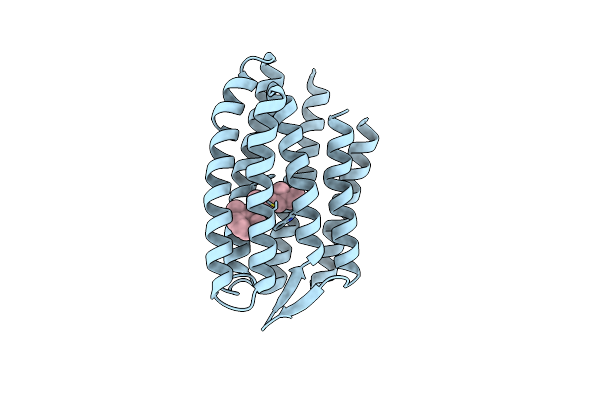 |
Continuously Illuminated Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL |
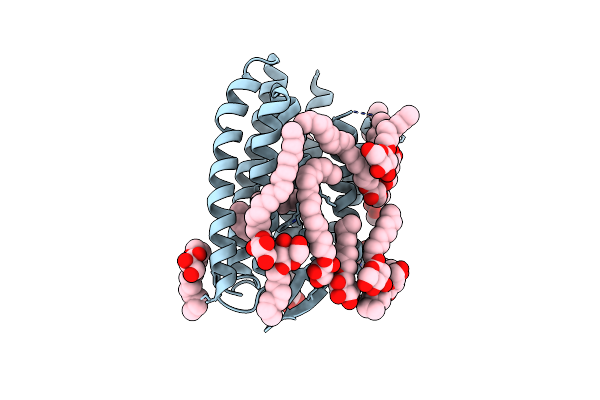 |
Continuous Dark State Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
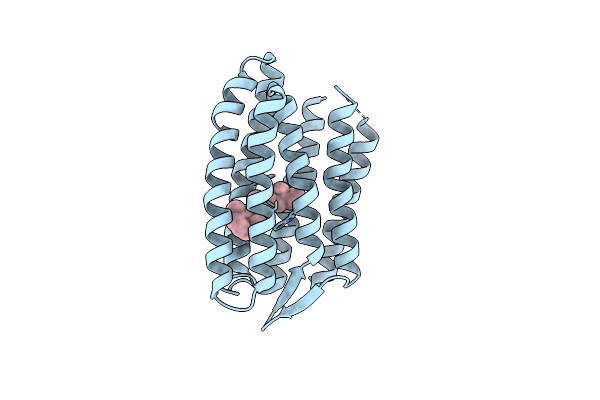 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 90-120 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 60-90 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography. 30-60 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 0-30 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 120-150 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Thaumatin Measured Via Serial Crystallography From A Kapton Hare-Chip (50 Micron)
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-05 Classification: SUGAR BINDING PROTEIN Ligands: TLA |
 |
Lysozyme Measured Via Serial Crystallography From A Kapton Hare-Chip (125 Micron)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-01-01 Classification: HYDROLASE Ligands: ACT, CL, NA |
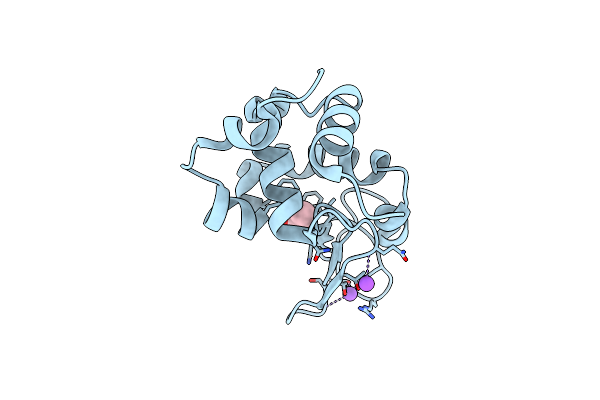 |
Lysozyme Measured Via Serial Crystallography From A Kapton Hare-Chip (50 Micron)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: HYDROLASE Ligands: ACT, CL, NA |
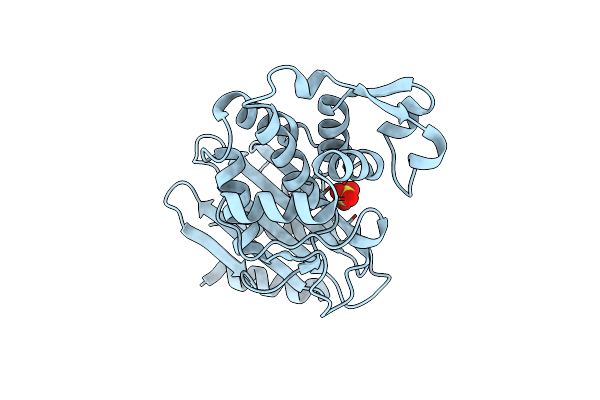 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: HYDROLASE Ligands: SO4 |
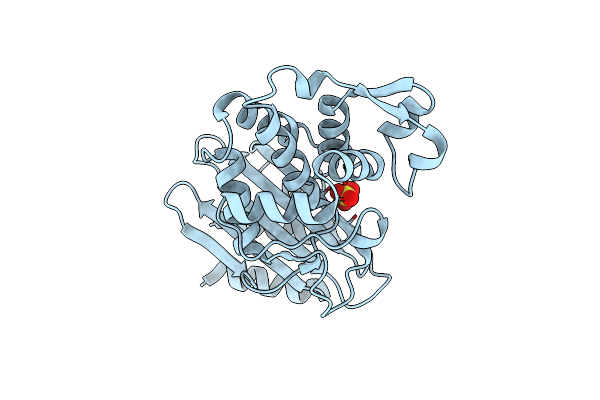 |
Ctx-M-14 Measured Via Serial Crystallography From A Kapton Hare-Chip (50 Micron)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-12-04 Classification: HYDROLASE Ligands: SO4 |
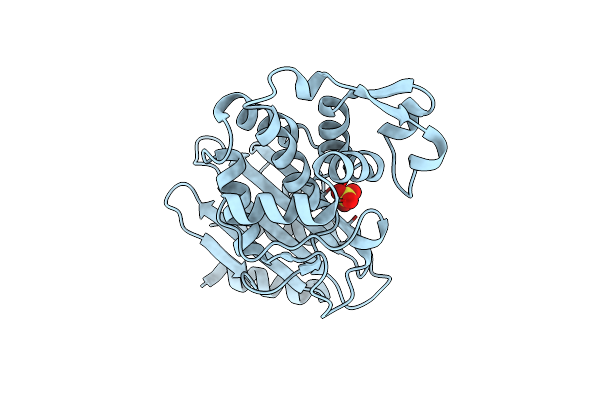 |
Ctx-M-14 Measured Via Serial Crystallography From A Kapton Hare-Chip (125 Micron)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-12-04 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: SUGAR BINDING PROTEIN Ligands: TLA |
 |
Thaumatin Measured Via Serial Crystallography From A Kapton Hare-Chip (125 Micron)
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-04 Classification: SUGAR BINDING PROTEIN Ligands: TAR |

