Search Count: 23
 |
Organism: Candidatus odinarchaeota
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: CELL CYCLE Ligands: G2P |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: DNA BINDING PROTEIN Ligands: GNP, MG |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-09-28 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, MES, PEG, CA, EDO |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-09-21 Classification: ANTIBIOTIC Ligands: NAP, GOL, CA, ACT, HMG, MEV |
 |
Targeting Enterococcus Faecalis Hmg-Coa Reductase With A Novel Non-Statin Inhibitor
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2022-09-21 Classification: ANTIBIOTIC Ligands: YPV, SO4, CA |
 |
Mycobacterium Tuberculosis Enolase Mutant - E204A Complex With Phosphoenolpyruvate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: PEP, PEG, EDO, ACT, MG |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN Ligands: PEP, MG |
 |
Mycobacterium Tuberculosis Enolase In Complex With Alternate 2-Phosphoglycerate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-01-26 Classification: LYASE Ligands: EDO, ACT, MG, 2PG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-01 Classification: LYASE Ligands: PEP, PEG, EDO, ACT, MG, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-07-28 Classification: LYASE Ligands: MG, 2PG, GOL, ACT, PEG, CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-07-21 Classification: LYASE Ligands: MG, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-11-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: EXF, SO4 |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: EDO, 2PG, ACT, MG |
 |
Crystal Structure Of Interleukin-1 Receptor-Associated Kinase 4 (Irak4-Wt) Complex With A Nicotinamide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-06-24 Classification: TRANSFERASE/TRANSFERASE Inhibitor Ligands: R7S, SO4 |
 |
Crystal Structure Of N-Acetylglucosamine-6-Phosphate Deacetylase From Pasteurella Multocida
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-03-04 Classification: METAL BINDING PROTEIN Ligands: ZN, PO4, EDO, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: FE, 1PE, PGE, PEG, EDO, P6G |
 |
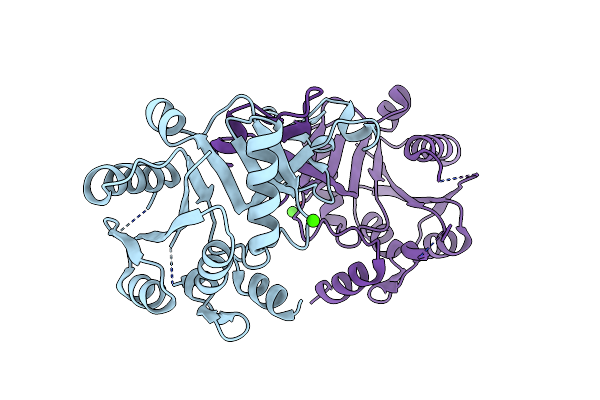
Crystal Structure Of Cmp-N-Acetylneuraminate Synthetase From Vibrio Cholerae In Complex With Cdp And Mg2+.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: SUGAR BINDING PROTEIN Ligands: CDP, PGE, EDO, CA, MG, PG4 |
 |
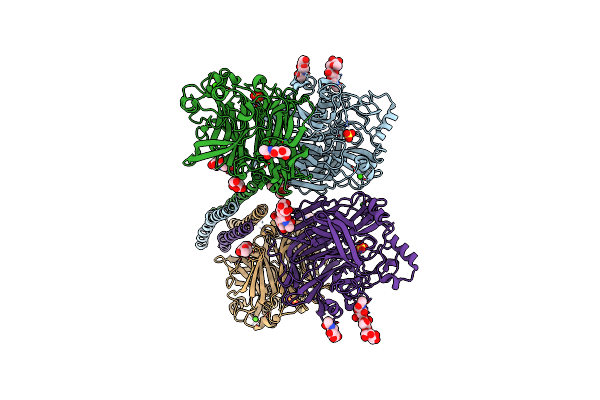
Crystal Structure Of The Apo Form Of Cmp-N-Acetylneuraminate Synthetase From Vibrio Cholerae
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-07-03 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Structure Of The Parainfluenza Virus 5 (Piv5) Hemagglutinin-Neuraminidase (Hn) Ectodomain
Organism: Simian virus 5
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-09-04 Classification: VIRAL PROTEIN Ligands: CA, SO4, NAG |

