Search Count: 156
 |
Structure Of Meiothermus Ruber Mrub_1259 Lov Domain With N- And C-Terminal Alpha Helices (Mrlove)
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Human Pigment Epithelium-Derived Factor With Zinc Ions Crystallized In P2(1)2(1)2(1) Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: ZN, SO4, PG0 |
 |
Human Pigment Epithelium-Derived Factor With Zinc Ion Crystallized In P22(1)2(1) Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: ZN, PG0 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-06-04 Classification: DE NOVO PROTEIN Ligands: RET, SO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Resolution:2.94 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Resolution:2.43 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Resolution:2.87 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA, RET |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The Ground State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET, LFA |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The M State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET |
 |
Organism: Subtercola endophyticus
Method: ELECTRON MICROSCOPY Resolution:2.44 Å Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
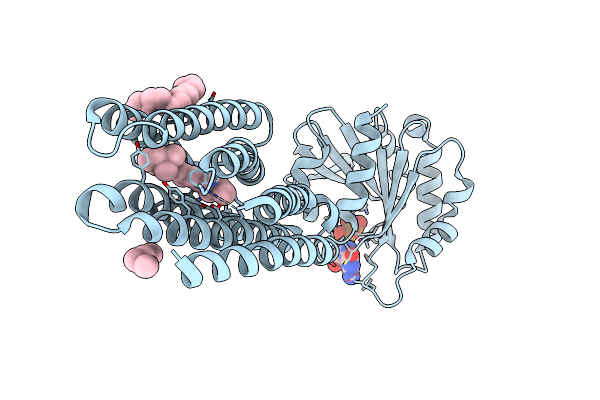 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
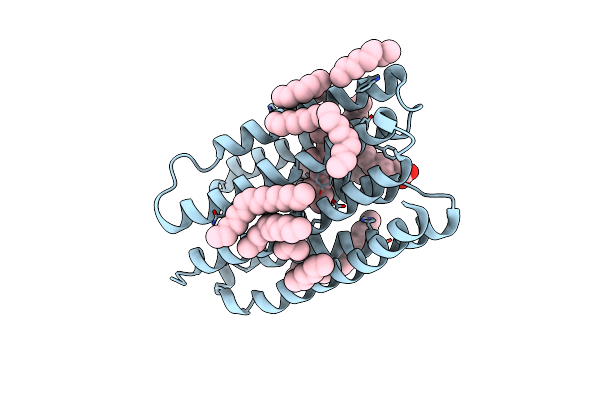 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
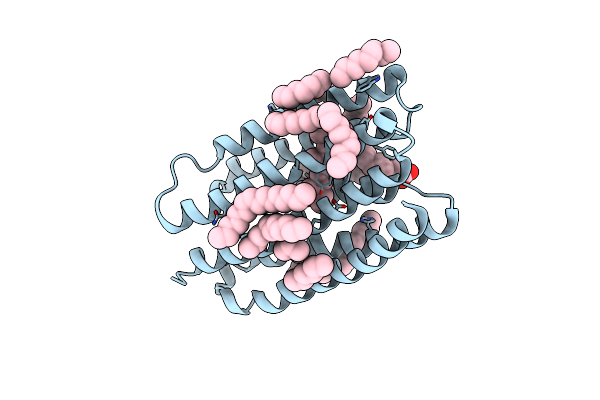 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |

