Search Count: 21
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: ISOMERASE Ligands: GDP, MG |
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: ISOMERASE |
 |
Meab In Complex With The Cobalamin-Binding Domain Of Its Target Mutase With Gmppcp Bound
Organism: Methylorubrum extorquens am1
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2023-03-01 Classification: CHAPERONE Ligands: GCP, MG, GOL |
 |
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: NAG, EDO, ACT, GOL, K |
 |
Gh3 Exo-Beta-Xylosidase (Xlnd) In Complex With Xylobiose Aziridine Activity Based Probe
Organism: Emericella nidulans (strain fgsc a4 / atcc 38163 / cbs 112.46 / nrrl 194 / m139)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: NAG, HLB, EDO |
 |
Organism: Aspergillus aculeatus atcc 16872
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: NAG, EDO, K |
 |
Organism: Aspergillus aculeatus atcc 16872
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: NAG, EDO, K, HQ8, XYP |
 |
Crystal Structure Of Aspergillus Niger Gh11 Endoxylanase Xyna In Complex With Xylobiose Epoxide Activity Based Probe
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: MES, SO4, HZB |
 |
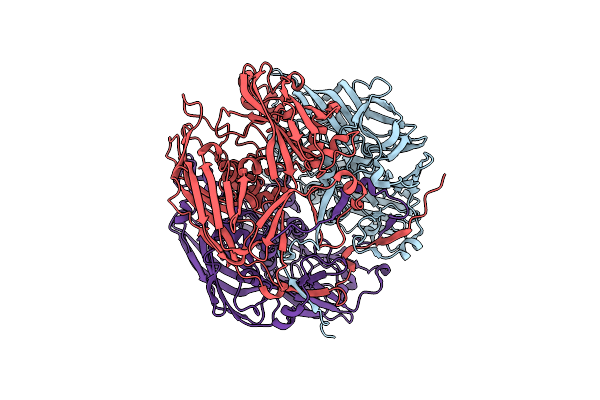
Organism: Cafeteriavirus-dependent mavirus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-07-04 Classification: VIRAL PROTEIN |
 |
Organism: Cafeteriavirus-dependent mavirus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-07-04 Classification: VIRAL PROTEIN |
 |
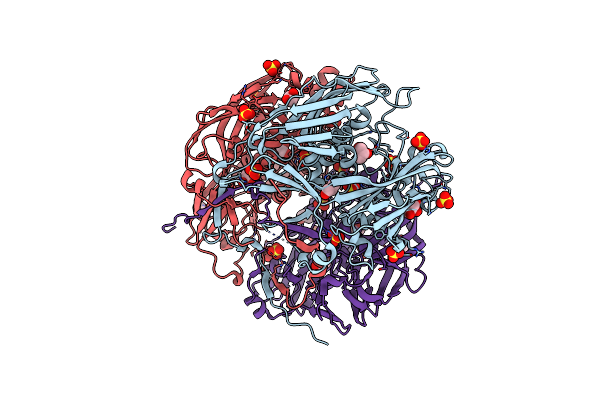
Crystal Structure Of Semet-Labeled Mavirus Major Capsid Protein Lacking The C-Terminal Domain
Organism: Cafeteriavirus-dependent mavirus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-07-04 Classification: VIRAL PROTEIN |
 |
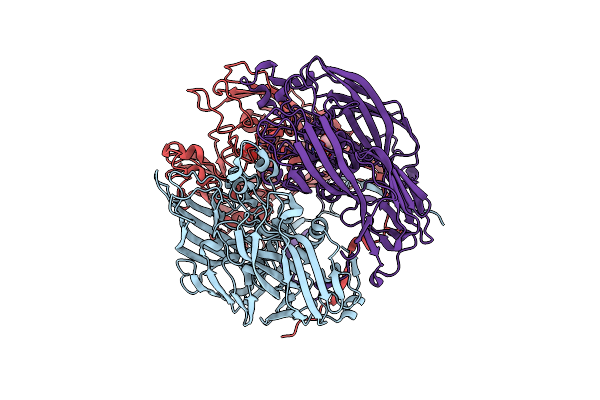
Crystal Structure Of Mavirus Major Capsid Protein Lacking The C-Terminal Domain
Organism: Cafeteriavirus-dependent mavirus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-07-04 Classification: VIRAL PROTEIN Ligands: SO4, GOL |
 |
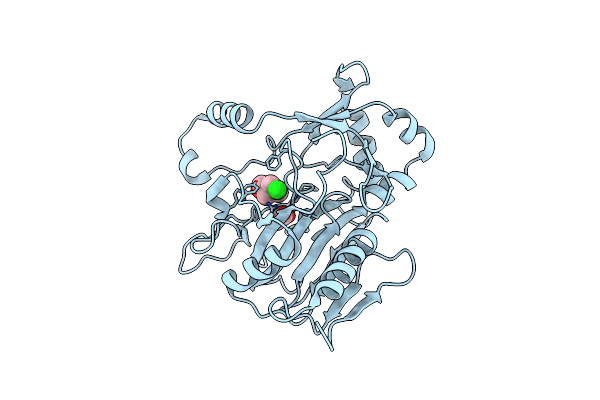
Organism: Cafeteriavirus-dependent mavirus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-04 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-06-27 Classification: OXIDOREDUCTASE Ligands: ACT, CL, ZN, CHT |
 |
Streptomyces Albus Hepd With Substrate 2-Hydroxyethylphosphonate (2-Hep) And Fe(Ii) Bound
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: FE, 2HE, GOL |
 |
Organism: Nitrosopumilus maritimus scm1
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Crystal Structure Of Mpns With Substrate 2-Hydroxyethylphosphonate (2-Hep) And Fe(Ii) Bound
Organism: Nitrosopumilus maritimus (strain scm1)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: FE, 2HE, FMT |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isobutyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, CO6, GDP, MG |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate N-Butyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, BCO, GDP, MG |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isovaleryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, IVC, GDP, MG |

