Search Count: 11
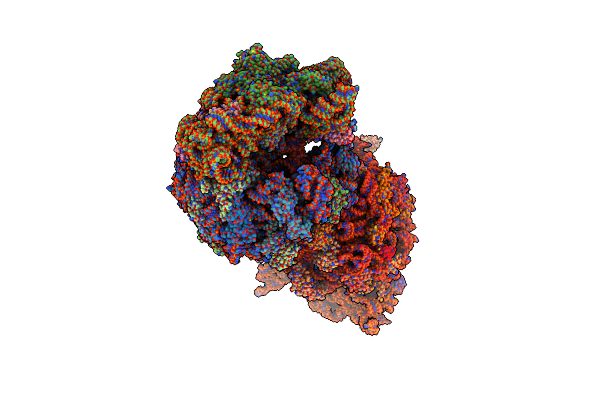 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Bac7-001, Mrna, And Deacylated P-Site Trna At 3.00A Resolution
Organism: Escherichia virus t4, Bos taurus, Thermus thermophilus hb8, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-08-26 Classification: RIBOSOME Ligands: MG, ZN, SF4, K |
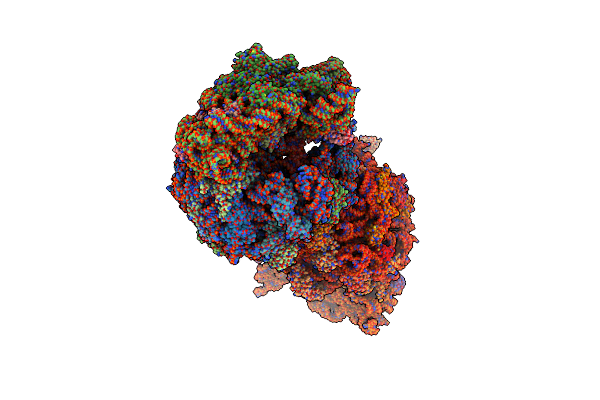 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Bac7-002, Mrna, And Deacylated P-Site Trna At 3.05A Resolution
Organism: Escherichia virus t4, Bos taurus, Thermus thermophilus hb8, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2020-08-26 Classification: RIBOSOME Ligands: MG, ZN, SF4, K |
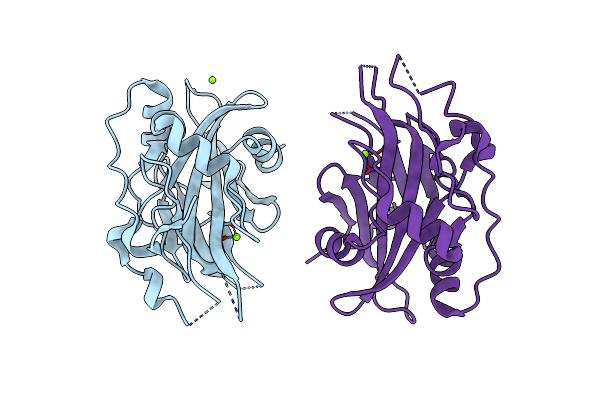 |
Organism: Pythium aphanidermatum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-08-28 Classification: TOXIN Ligands: MG |
 |
Organism: Hyaloperonospora arabidopsidis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-08-28 Classification: TOXIN Ligands: NAG, PO4 |
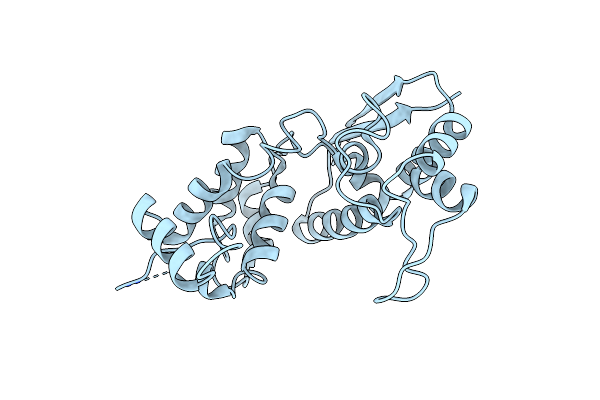 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-03-20 Classification: HYDROLASE Ligands: CL |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2019-03-20 Classification: HYDROLASE Ligands: PG4, CL, NA |
 |
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-02-08 Classification: CELL ADHESION Ligands: SO4, CL |
 |
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-02-08 Classification: HYDROLASE Ligands: CA, NA, CL, ZN, CIT |
 |
Development Of N-(Functionalized Benzoyl)-Homocycloleucyl-Glycinonitriles As Potent Cathepsin K Inhibitors.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2015-09-30 Classification: HYDROLASE Ligands: 3Y1, SO4 |
 |
Development Of N-(Functionalized Benzoyl)-Homocycloleucyl-Glycinonitriles As Potent Cathepsin K Inhibitors.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2015-09-23 Classification: HYDROLASE Ligands: 3XT, I37, SO4 |
 |
Development Of N-(Functionalized Benzoyl)-Homocycloleucyl-Glycinonitriles As Potent Cathepsin K Inhibitors.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2015-09-09 Classification: HYDROLASE Ligands: GOL, CL, K, PGO, 3Y2 |

