Search Count: 15
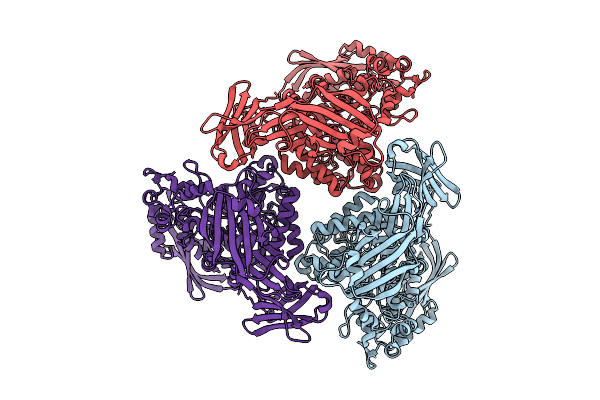 |
Active Conformation Of A Redox-Regulated Glycoside Hydrolase (Capgh2B) From The Gh2 Family
Organism: Metagenome
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2025-11-12 Classification: HYDROLASE |
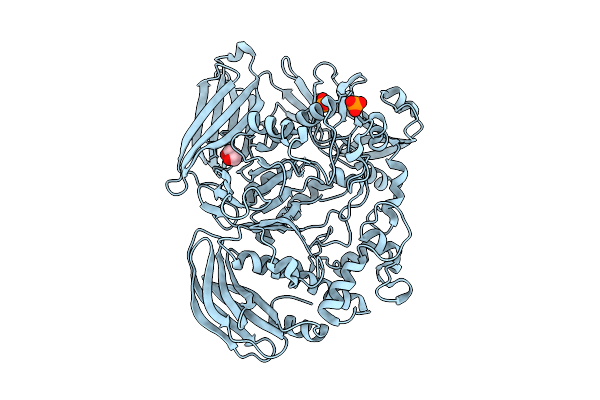 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
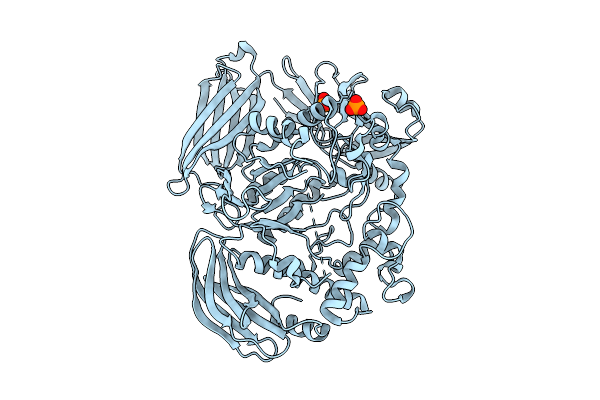 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group I213 At 2.6 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4 |
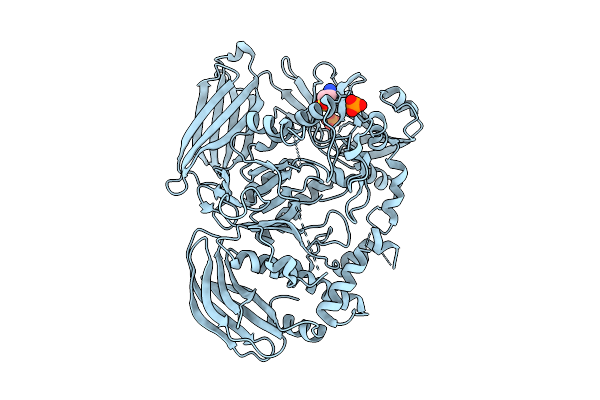 |
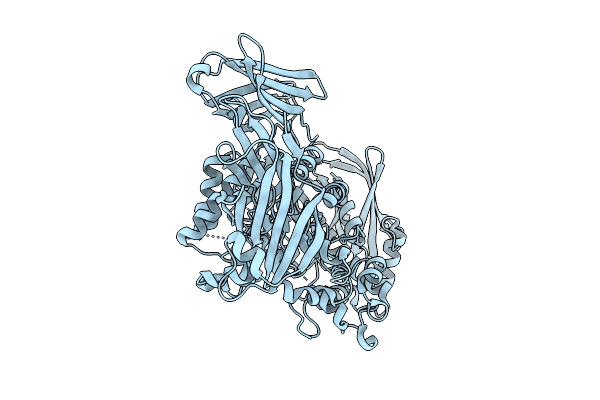
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.75 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, TAU |
 |
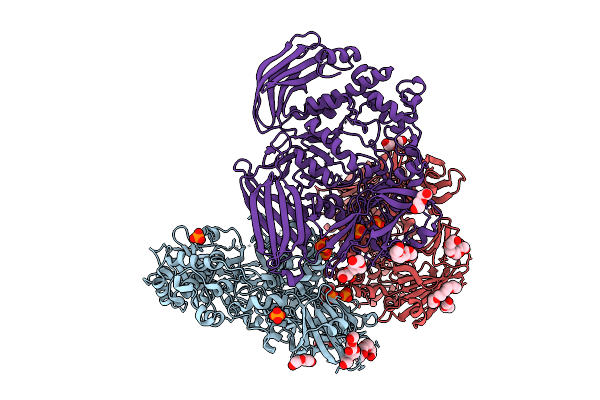
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group R3 At 2.45 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-11-12 Classification: HYDROLASE |
 |
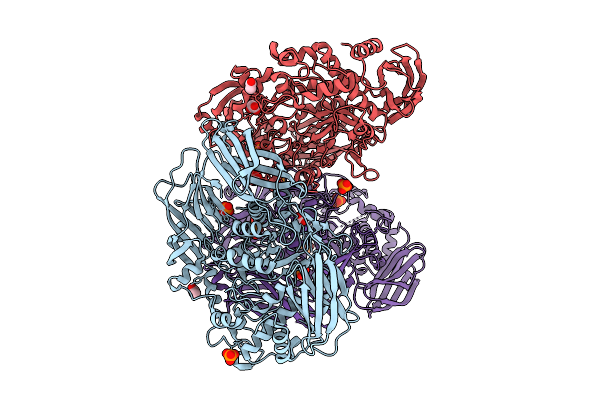
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I212121 At 2.65 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, PEG, PG4 |
 |
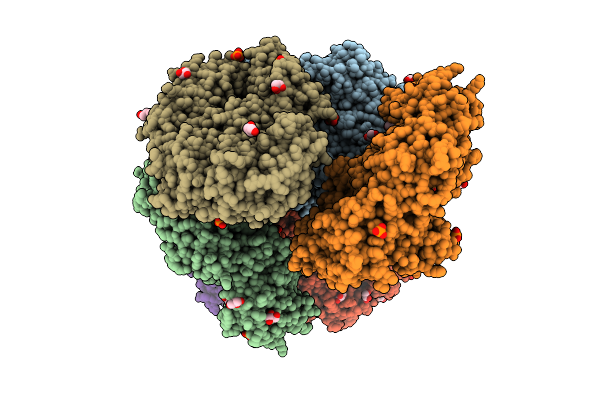
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P3121 At 3.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, ACT, MLI |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.40 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.15 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL, ACT |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P1 At 2.25 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E465A Mutant) From The Gh2 Family In The Space Group P1 At 3.1 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Cryo-Em Structure Of Bl_Man38A Nucleophile Mutant In Complex With Mannose At 2.7 A
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN, MAN |

