Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
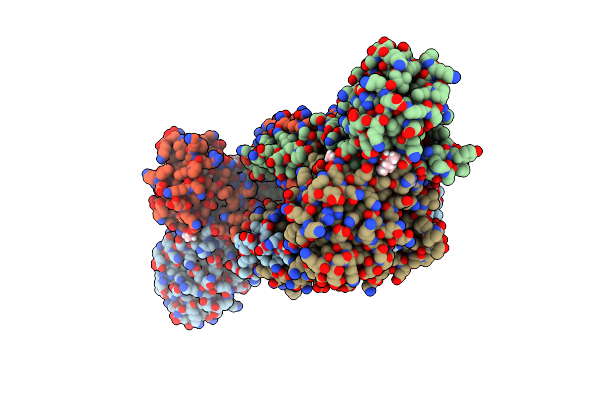 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL, MPD |
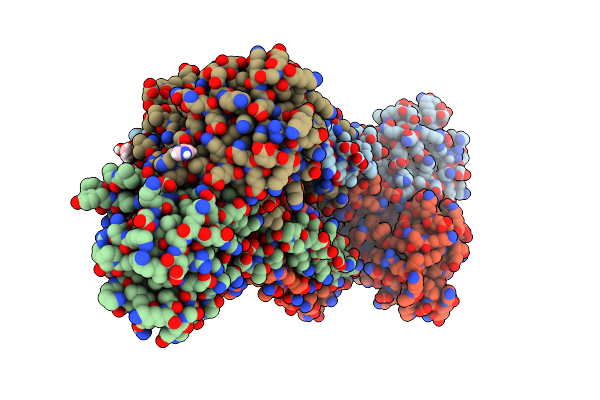 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL, IMD |
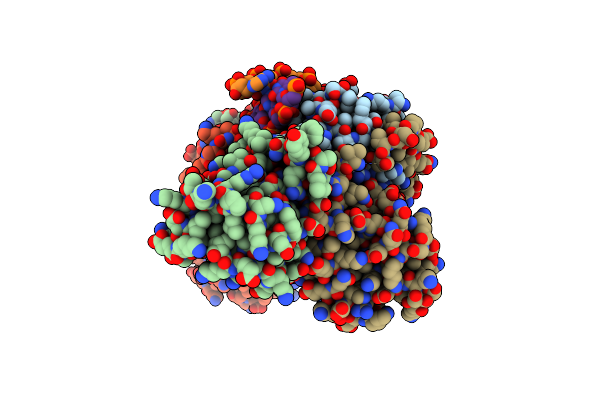 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL |
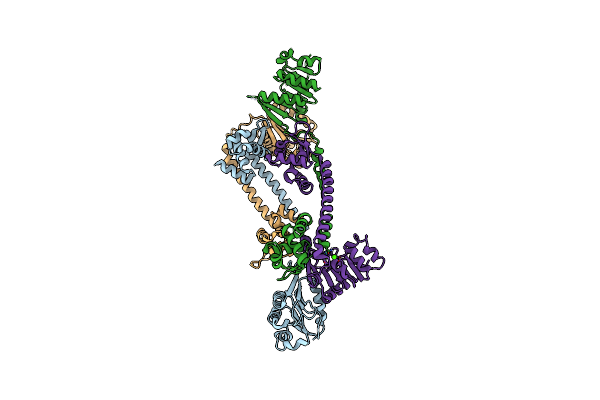 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: CA |
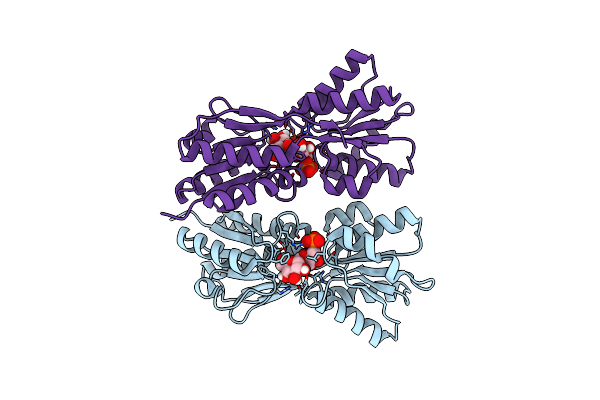 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-02-11 Classification: GENE REGULATION |
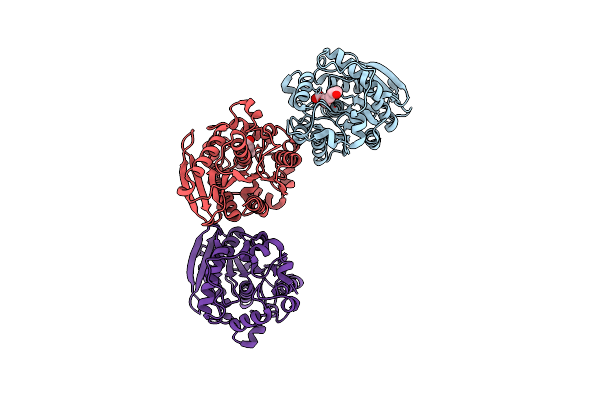 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: PG4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: PGE, IMD, 1PE |
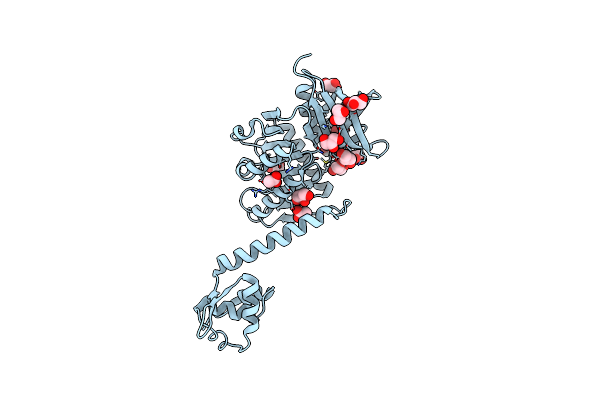 |
The Three-Dimensional Structure Of Trmb, A Global Transcriptional Regulator Of The Hyperthermophilic Archaeon Pyrococcus Furiosus In Complex With Sucrose
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2012-05-30 Classification: TRANSCRIPTION Ligands: ACT, GOL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: PEG, NA |
 |
Crystal Structure Of Glycogen Debranching Enzyme Glgx From Escherichia Coli K-12
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2010-09-01 Classification: HYDROLASE Ligands: SO4 |
 |
X-Ray Structure Of Glucose/Galactose Receptor From Salmonella Typhimurium In Complex With (2R)-Glyceryl-Beta-D-Galactopyranoside
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2009-04-14 Classification: SUGAR BINDING PROTEIN Ligands: RGG, CA, SCN, NA |
 |
Crystal Structure Of A Major Outer Membrane Protein From Thermus Thermophilus Hb27
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-12-23 Classification: UNKNOWN FUNCTION Ligands: CA, C8E |
 |
Solution Structure Of The Soluble Domain Of The Nfed Protein Yuaf From Bacillus Subtilis
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2008-08-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of The Sugar Binding Domain Of The Archaeal Transcriptional Regulator Trmb
Organism: Thermococcus litoralis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-02-21 Classification: TRANSCRIPTION Ligands: IMD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2005-06-14 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Thermococcus litoralis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-03-14 Classification: SUGAR BINDING PROTEIN Ligands: PT, CL |
 |
Organism: Thermococcus litoralis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-12-06 Classification: SUGAR BINDING PROTEIN Ligands: NH4, CL, NA, MG, POP, DIO |
 |
X-Ray Structure Of Maly From Escherichia Coli: A Pyridoxal-5'-Phosphate-Dependent Enzyme Acting As A Modulator In Mal Gene Expression
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2000-01-12 Classification: TRANSFERASE Ligands: PLP |