Search Count: 25
 |
Structure Of Human Cysteine Desulfurase Nfs1 With L-Propargylglycine Bound To Active Site Plp In Complex With Isd11, Acp1 And Iscu2
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-11 Classification: TRANSFERASE Ligands: EDO, PLP, LPH, GOL, PEG, PG4, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-04-06 Classification: METAL BINDING PROTEIN Ligands: GSH, EDO, MES, PEG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-01-19 Classification: METAL BINDING PROTEIN Ligands: EDO, CU1, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-11-17 Classification: METAL BINDING PROTEIN Ligands: GSH, EDO, FE2, GOL, PEG |
 |
Structure Of The (Niau)2 Complex With N-Terminal Mutation Of Iscu2 Y35D At 2.5 A Resolution
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-08-25 Classification: TRANSFERASE Ligands: PLP, EDO, GOL, PG4, PEG, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
N-Terminal Mutation Of Iscu2 (L35H36) Traps Nfs1 Cys Loop In The Active Site Of Iscu2 Without Metal Present. Structure Of Human Mitochondrial Complex Nfs1-Iscu2(L35H36)-Isd11 With E.Coli Acp1 At 1.9 A Resolution (Niau)2.
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-13 Classification: TRANSFERASE Ligands: PLP, PEG, GOL, EDO, PG4, P15, PGE, EDT, 8Q1, 1PE |
 |
Structure Of Human Mitochondrial Complex Nfs1-Iscu2-Isd11 With E.Coli Acp1 At 1.95 A Resolution (Niau)2. N-Terminal Mutation Of Iscu2 (L35) Traps Nfs1 Cys Loop In The Active Site Of Iscu2 Without Metal Present.
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-04-22 Classification: TRANSFERASE Ligands: PLP, EDO, GOL, PEG, PG4, ETE, PGE, MES, EDT, 8Q1, 1PE |
 |
Structure Of Human Mitochondrial Complex Nfs1-Iscu2 (Wt)-Isd11 With E.Coli Acp1 At 1.8 A Resolution (Niau)2
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-18 Classification: TRANSFERASE Ligands: PLP, GOL, EDO, PEG, PG4, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
Structure Of The Human Mitochondrial Desulfurase Complex Nfs1-Iscu2(M140I)-Isd11 With E.Coli Acp1 At 1.57 A Resolution Showing Flexibility Of N Terminal End Of Iscu2
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-11-20 Classification: TRANSFERASE Ligands: PLP, GOL, EDO, PEG, PG4, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
L. Pneumophila Effector Kinase Legk7 (Amp-Pnp Bound) In Complex With Human Mob1A
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: PG4, PEG, P6G, ZN, MN, ANP |
 |
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: PEG, P6G, PG4, ZN |
 |
Structure Of Lpne Effector Protein From Legionella Pneumophila (Sp. Philadelphia)
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-19 Classification: PROTEIN BINDING Ligands: NI, GOL, PEG |
 |
Crystal Structure Of The Human Mitochondrial Cysteine Desulfurase In Complex With Isd11 And Iron-Sulfur Cluster Scaffold Protein Iscu1, And E. Coli Acp1 Protein At 3.15A
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2017-11-15 Classification: TRANSFERASE Ligands: PLP, 8Q1 |
 |
Crystal Structure Of The Human Mitochondrial Cysteine Desulfurase With Active Cysteine Loop Within Iscu1 Active Site, Coordinating Zn Ion. Complexed With Human Isd11 And E. Coli Acp1 At 3.3A.
Organism: Homo sapiens, Escherichia coli o45:k1 (strain s88 / expec)
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2017-11-15 Classification: TRANSFERASE Ligands: PLP, 8Q1, ZN |
 |
Crystal Structure Of The Human Mitochondrial Cysteine Desulfurase In Complex With Isd11 And E. Coli Acp1 Protein At 2.75A
Organism: Homo sapiens, Escherichia coli o45:k1 (strain s88 / expec)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: PLP, 8Q1 |
 |
Yeast V-Atpase In Complex With Legionella Pneumophila Effector Sidk (Rotational State 3)
Organism: Legionella pneumophila subsp. pneumophila atcc 43290, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2017-06-28 Classification: HYDROLASE |
 |
Yeast V-Atpase In Complex With Legionella Pneumophila Effector Sidk (Rotational State 1)
Organism: Legionella pneumophila subsp. pneumophila atcc 43290, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2017-06-21 Classification: HYDROLASE |
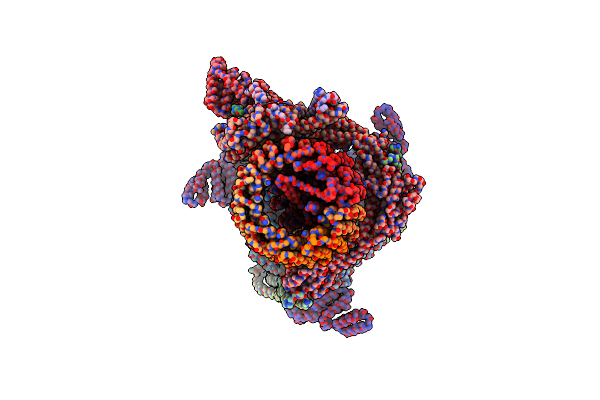 |
Yeast V-Atpase In Complex With Legionella Pneumophila Effector Sidk (Rotational State 2)
Organism: Legionella pneumophila subsp. pneumophila atcc 43290, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2017-06-21 Classification: HYDROLASE |
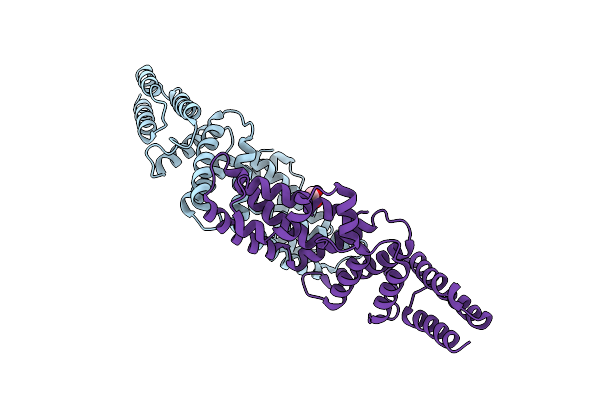 |
Structure Of The Effector Protein Sidk (Lpg0968) From Legionella Pneumophila (Domain-Swapped Dimer)
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-05-10 Classification: PROTEIN BINDING Ligands: GOL |
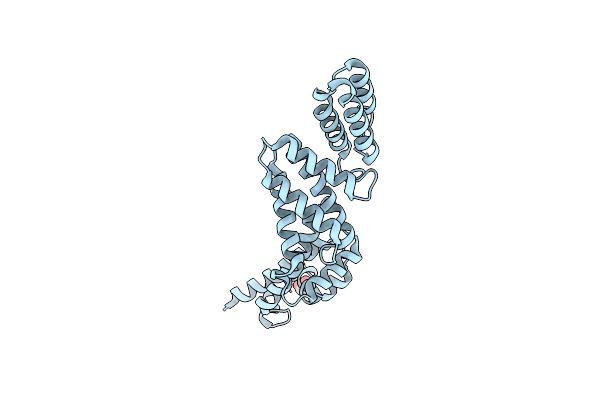 |
Structure Of The Effector Protein Sidk (Lpg0968) From Legionella Pneumophila
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-05-10 Classification: PROTEIN BINDING Ligands: GOL |

