Search Count: 18
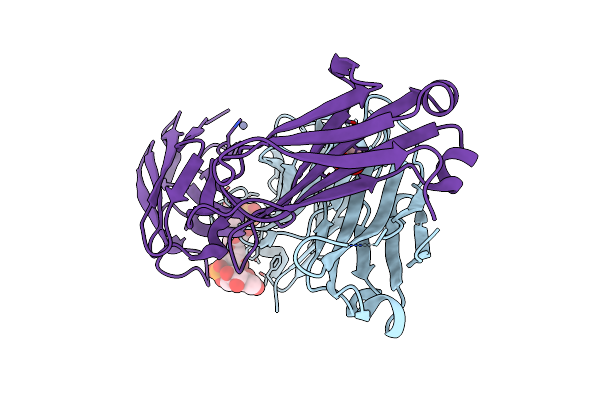 |
Crystal Structure Of Haemophilus Influenzae Type B (Hib) Dp2 Oligosaccharide Bound To Fab Ca4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-05-15 Classification: IMMUNE SYSTEM Ligands: A1H1A, ZN, EDO |
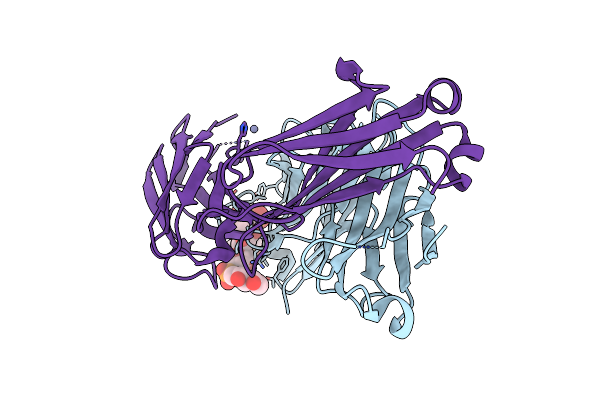 |
Crystal Structure Of Haemophilus Influenzae Type B (Hib) Dp3 Oligosaccharide Bound To Fab Ca4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-05-15 Classification: IMMUNE SYSTEM Ligands: A1H09, ZN |
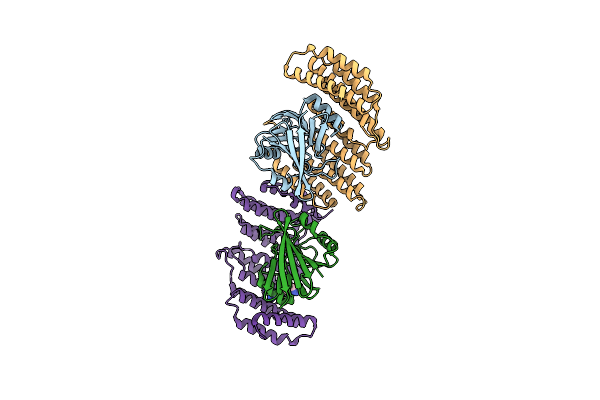 |
Crystal Structure Of The Retromer Complex Vps29/Vps35 With The Ligand Bis-1,3-Phenyl Guanylhydrazone, 2A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-03-27 Classification: PROTEIN TRANSPORT Ligands: XFZ |
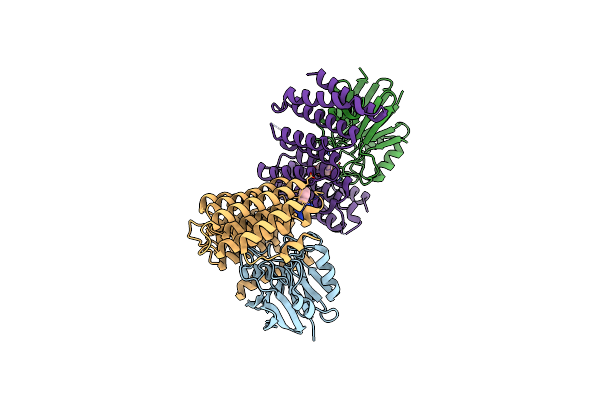 |
Crystal Structure Of The Retromer Complex Vps29/Vps35 With The Ligand Bis-1,3-Phenyl Guanylhydrazone, 2A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-03-27 Classification: PROTEIN TRANSPORT Ligands: XFZ, TRS |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With The 5-[(Morpholin-4-Yl)Methyl]Quinolin-8-Ol Inhibitor
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: JM3, NAG, MAN |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With Udp-2-Deoxy-2-Fluoro-D-Glucose
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: NAG, U2F, CA, PDO |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum (Apo Form)
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of Diphtheria Toxin Mutant Crm197 With A Disulphide Bond Replaced By A Cys-Acetone-Cys Bridge
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: 4Y8, EDO, PGE, ACT |
 |
Crystal Structure Of The Pathological N184K Variant Of Calcium-Free Human Gelsolin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: CL, GOL, SO4, TRS |
 |
Crystal Structure Of The Pathological G167R Variant Of Calcium-Free Human Gelsolin,
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL |
 |
Crystal Structure Of The Pathological D187N Variant Of Calcium-Free Human Gelsolin.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-08-28 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-11-08 Classification: STRUCTURAL PROTEIN Ligands: CA, FLC, ACT, NA, GOL |
 |
Crystal Structure Of Group B Streptococcus Type Iii Dp2 Oligosaccharide Bound To Fab Nvs-1-19-5
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2017-05-03 Classification: IMMUNE SYSTEM Ligands: EDO, 7GW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-05 Classification: STRUCTURAL PROTEIN Ligands: CA, SO4, PEG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2016-10-05 Classification: STRUCTURAL PROTEIN Ligands: CA, GOL, CL, ACT |
 |
Crystal Structure Of An Engineered Oxa-10 Variant With Carbapenemase Activity, Oxa-10Loop24
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-11-02 Classification: HYDROLASE Ligands: SO4, EDO |
 |
Crystal Structure Of A Rationally Designed Oxa-10 Variant Showing Carbapenemase Activity, Oxa-10Loop48
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-02 Classification: HYDROLASE Ligands: SO4, EDO, CO2 |

