Search Count: 9
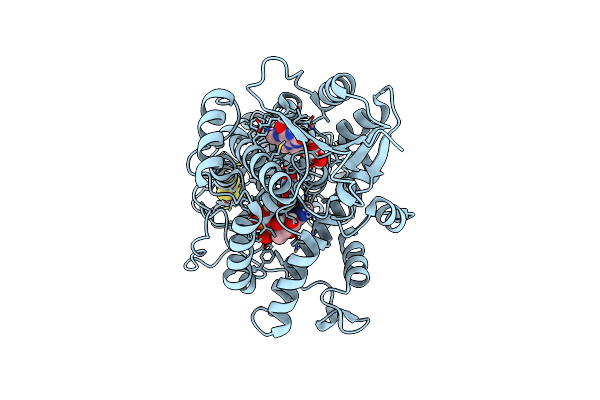 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Oxidizd State - Singal Crystal / Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
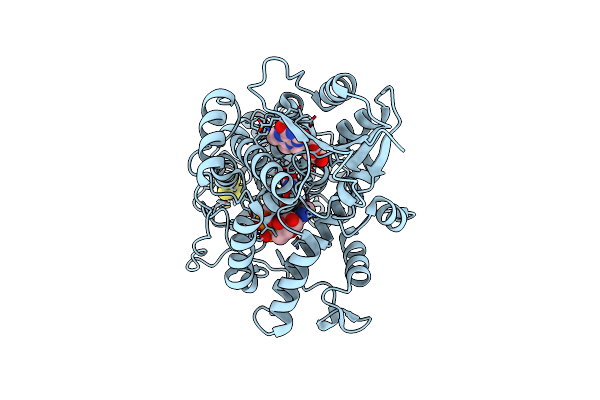 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Oxidized State At Room Temperture-Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
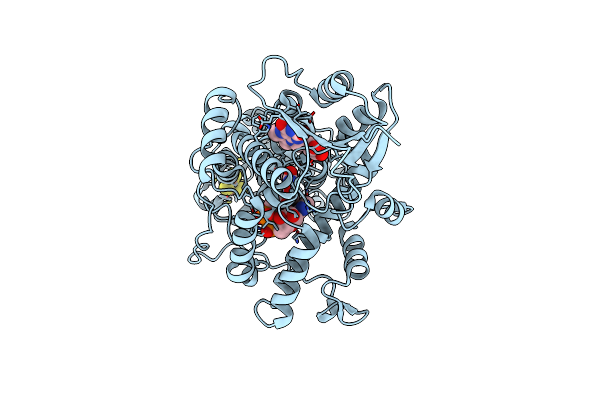 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Dark Adapted And Oxidized State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
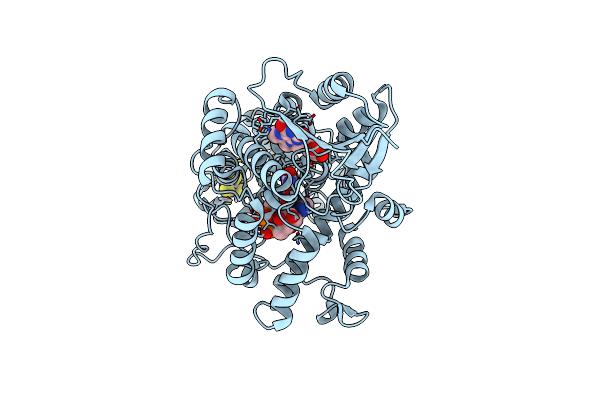 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Fully Reduced State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Iron Sulfur Cluster Oxidized State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus With K48A Mutation In Its Dark Adapted And Oxidized State Determined By Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4, GOL, CL, SO4, MG |
 |
X-Ray Structure Of Perm1, A Circularly Permuted Mutant Of The Sweet Protein Mnei
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: MPD, ACT, MG, P6G |
 |
X-Ray Structure Of Perm3, A Circularly Permuted Mutant Of The Sweet Protein Mnei
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN |
 |
X-Ray Structure Of Perm2, A Circularly Permuted Mutant Of The Sweet Protein Mnei
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: ACY |

