Search Count: 15
 |
Crystal Structure Of Meganuclease I-Smami Bound To Uncleaveable Dna With A Ttct Central Four
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-30 Classification: hydrolase/dna Ligands: CA, GOL |
 |
Crystal Structure Of Meganuclease I-Smami Bound To Uncleaveable Dna With A Ttgt Central Four
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-30 Classification: hydrolase/dna Ligands: CA, GOL |
 |
Crystal Structure Of Laglidadg Meganuclease I-Panmi With Coordinated Calcium Ions
Organism: Podospora anserina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-30 Classification: HYDROLASE/DNA Ligands: CA, GOL |
 |
Organism: Cryphonectria parasitica, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2016-03-23 Classification: hydrolase/dna Ligands: CA, EDO |
 |
Organism: Gremmeniella abietina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2016-03-23 Classification: hydrolase/dna Ligands: CA |
 |
Organism: Grosmannia penicillata, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-03-09 Classification: hydrolase/dna Ligands: CA |
 |
Computational Design Of An Unnatural Amino Acid Metalloprotein With Atomic Level Accuracy
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: METAL BINDING PROTEIN Ligands: SO4, CO |
 |
Computational Design Of An Unnatural Amino Acid Metalloprotein With Atomic Level Accuracy
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-08-21 Classification: METAL BINDING PROTEIN Ligands: NI, SO4 |
 |
Crystal Structure Of A Putative, De Novo Designed Unnatural Amino Acid Dependent Metalloprotein, Northeast Structural Genomics Consortium Target Or61
Organism: Micromonospora viridifaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-03-27 Classification: Structural Genomics, Unknown Function Ligands: GOL, ARS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: ZN |
 |
The Structure And Dna Recognition Of A Bifunctional Homing Endonuclease And Group I Intron Splicing Factor
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-01-20 Classification: HYDROLASE/DNA Ligands: MG |
 |
Isocitrate Dehydrogenase From E.Coli (Mutant K230M), Steady-State Intermediate Complex Determined By Laue Crystallography
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (NAD(A)-CHOH(D)) Ligands: MG, OXS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (NAD(A)-CHOH(D)) |
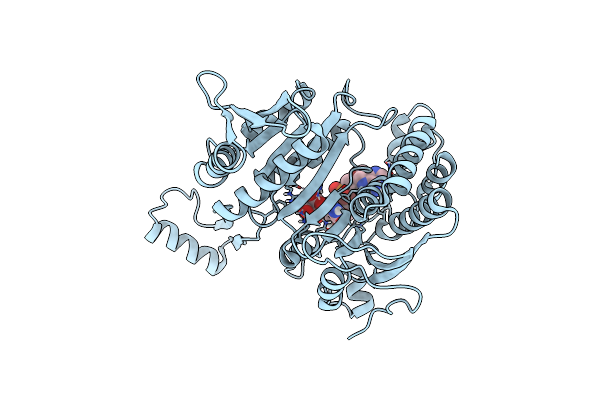 |
Isocitrate Dehydrogenase Y160F Mutant Steady-State Intermediate Complex (Laue Determination)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (NAD(A)-CHOH(D)) Ligands: MG, ICT, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (NAD(A)-CHOH(D)) |

