Search Count: 25
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
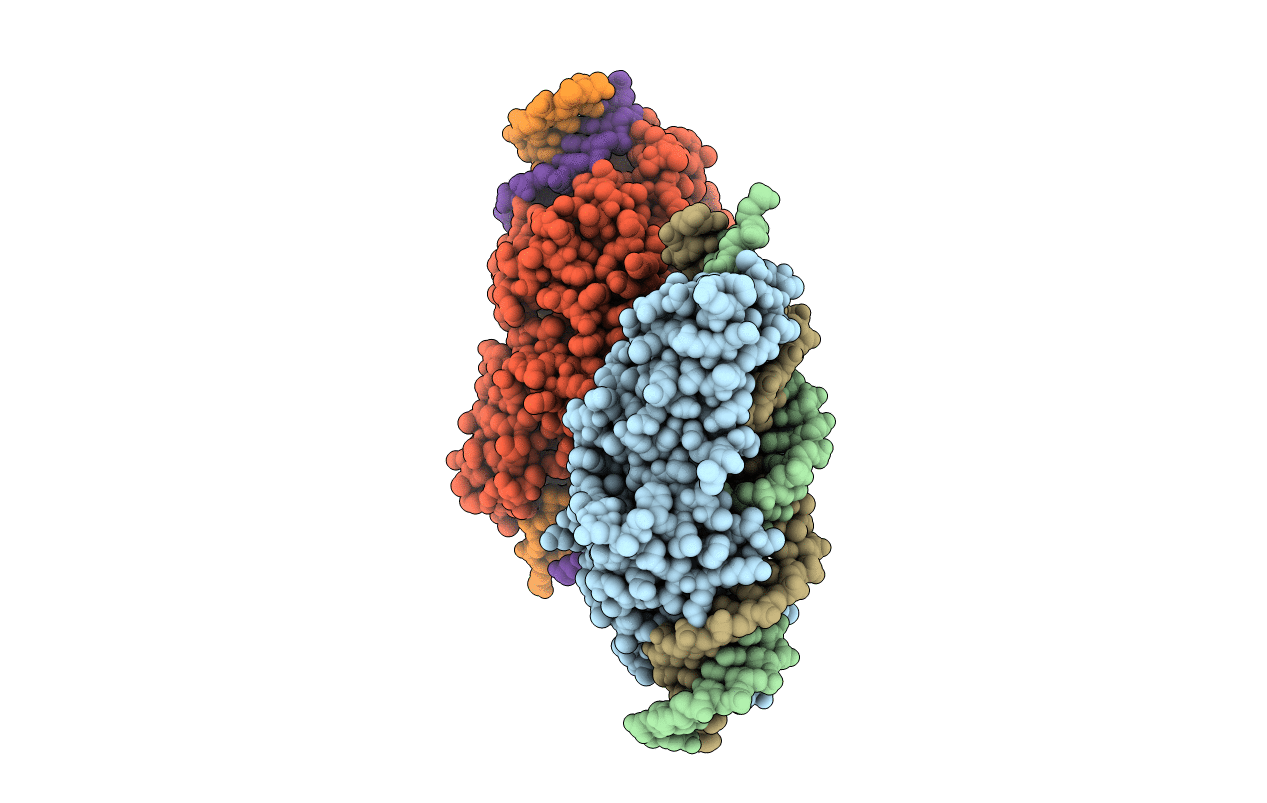
Crystal Structure Of Meganuclease I-Smami Bound To Uncleaveable Dna With A Ttct Central Four
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-30 Classification: hydrolase/dna Ligands: CA, GOL |
 |
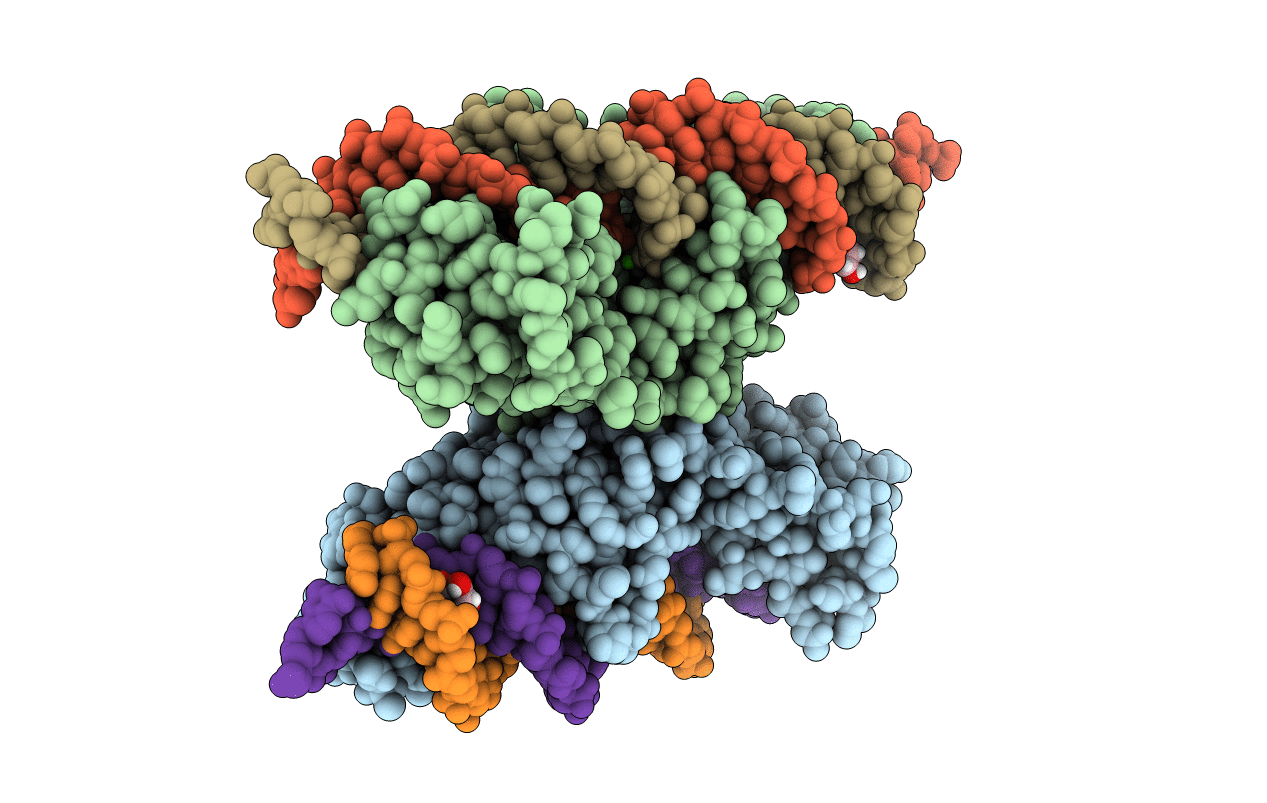
Crystal Structure Of Meganuclease I-Smami Bound To Uncleaveable Dna With A Ttgt Central Four
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-30 Classification: hydrolase/dna Ligands: CA, GOL |
 |
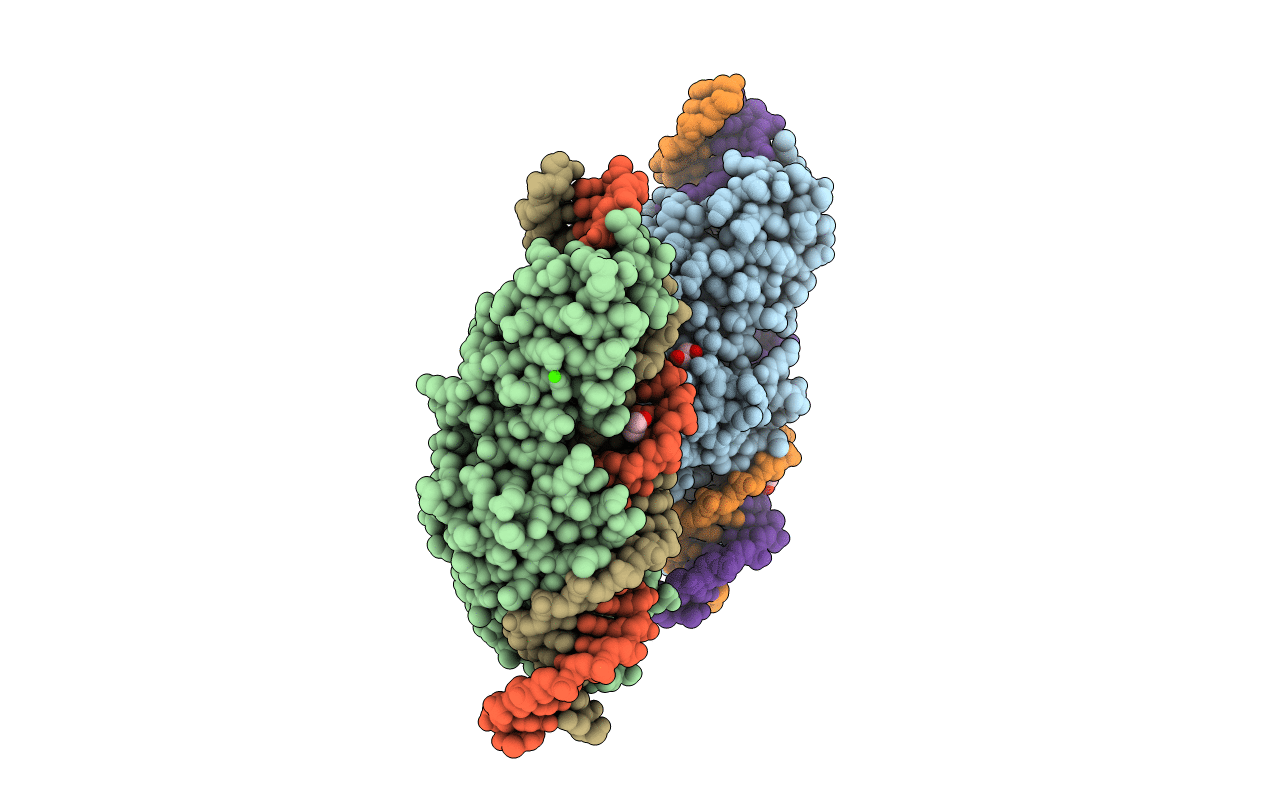
Crystal Structure Of Laglidadg Meganuclease I-Panmi With Coordinated Calcium Ions
Organism: Podospora anserina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-30 Classification: HYDROLASE/DNA Ligands: CA, GOL |
 |
Organism: Cryphonectria parasitica, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2016-03-23 Classification: hydrolase/dna Ligands: CA, EDO |
 |
Organism: Gremmeniella abietina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2016-03-23 Classification: hydrolase/dna Ligands: CA |
 |
Organism: Grosmannia penicillata, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-03-09 Classification: hydrolase/dna Ligands: CA |
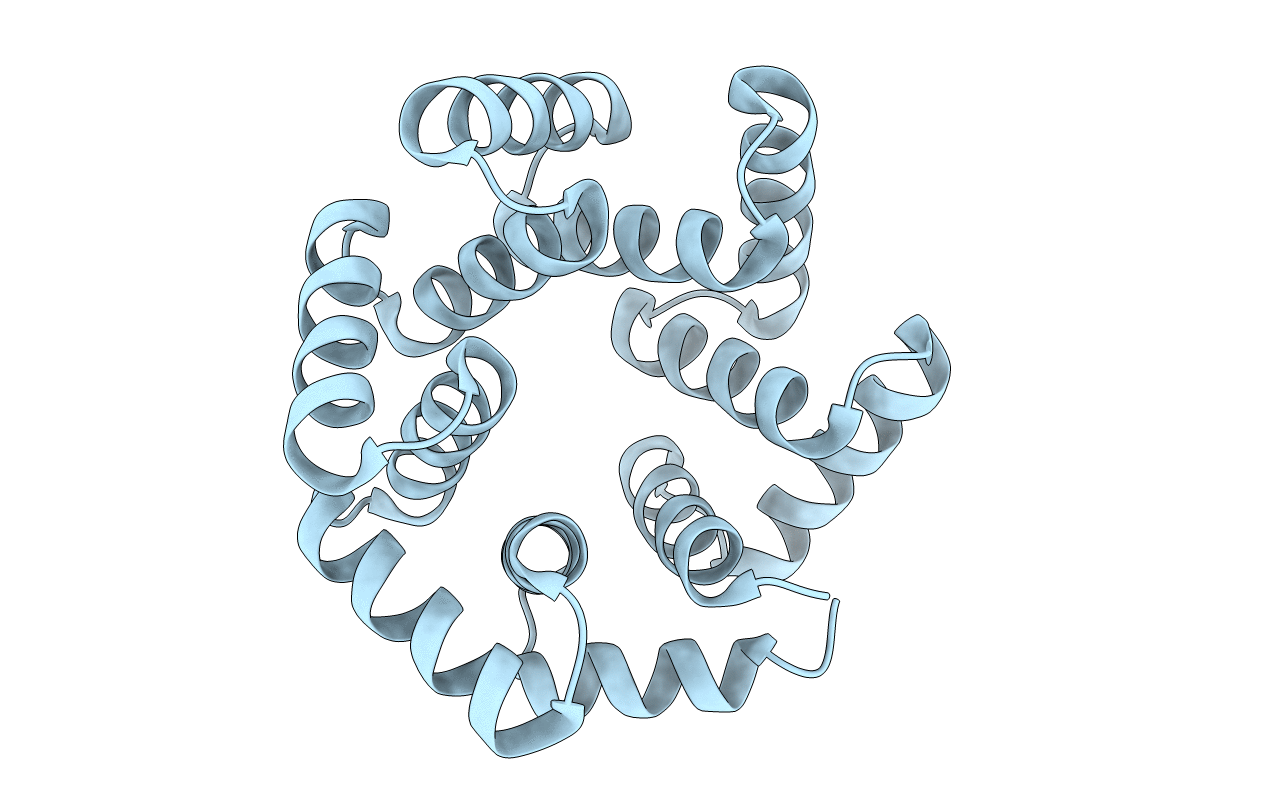 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Computationally Designed Left-Handed Alpha/Alpha Toroid With 9 Repeats; Two Linked Rings Of 12 Repeats Each Structure
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Computationally Designed Left-Handed Alpha/Alpha Toroid With 3 Repeats In Space Group P212121
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Computationally Designed Left-Handed Alpha/Alpha Toroid With 3 Repeats In Space Group P43212
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-16 Classification: DE NOVO PROTEIN |
 |
Computational Design Of An Unnatural Amino Acid Metalloprotein With Atomic Level Accuracy
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: METAL BINDING PROTEIN Ligands: SO4, CO |
 |
Computational Design Of An Unnatural Amino Acid Metalloprotein With Atomic Level Accuracy
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-08-21 Classification: METAL BINDING PROTEIN Ligands: NI, SO4 |
 |
Crystal Structure Of A Putative, De Novo Designed Unnatural Amino Acid Dependent Metalloprotein, Northeast Structural Genomics Consortium Target Or61
Organism: Micromonospora viridifaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-03-27 Classification: Structural Genomics, Unknown Function Ligands: GOL, ARS |
 |
Structural Analyses Of Covalent Enzyme-Substrate Analogue Complexes Reveal Strengths And Limitations Of De Novo Enzyme Design
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-11-23 Classification: LYASE/LYASE INHIBITOR Ligands: 3NK, SO4 |
 |
Robust Computational Design, Optimization, And Structural Characterization Of Retroaldol Enzymes
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-06-29 Classification: LYASE Ligands: SO4 |
 |
Robust Computational Design, Optimization, And Structural Characterization Of Retroaldol Enzymes
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-06-29 Classification: LYASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: ZN |

