Search Count: 24
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-05-22 Classification: TRANSPORT PROTEIN Ligands: B12, GSH, CL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-05-22 Classification: TRANSPORT PROTEIN Ligands: B12, NO3, CL, NA |
 |
Molecular Basis Of Zp3/Zp1 Heteropolymerization: Crystal Structure Of A Native Vertebrate Egg Coat Filament
Organism: Oryzias latipes
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-03-13 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of The Complete N-Terminal Region Of Human Zp2 (Hzp2-N1N2N3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Crystal Structure Of Tetrameric Collagenase-Cleaved Xenopus Zp2-N2N3 (Cleaved Xzp2-N2N3)
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG, NO3, BCN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Molecular Basis Of Zp3/Zp1 Heteropolymerization: Crystal Structure Of A Native Vertebrate Egg Coat Filament Fragment
Organism: Oryzias latipes
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2024-03-13 Classification: STRUCTURAL PROTEIN Ligands: YB |
 |
Cryo-Em Of Native Human Uromodulin (Umod)/Tamm-Horsfall Protein (Thp) Filament.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Cryo-Em Of Elastase-Treated Human Uromodulin (Umod)/Tamm-Horsfall Protein (Thp) Filament
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-06-07 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The Orphan Region Of Human Endoglin/Cd105 In Complex With Bmp9
Organism: Escherichia coli k12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.45 Å Release Date: 2017-06-07 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Escherichia coli k-12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2017-06-07 Classification: SIGNALING PROTEIN Ligands: NAG, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-06-07 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The Polymerization Region Of Human Uromodulin/Tamm-Horsfall Protein
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-01-27 Classification: STRUCTURAL PROTEIN Ligands: NAG, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-01-27 Classification: CELL ADHESION Ligands: ACT |
 |
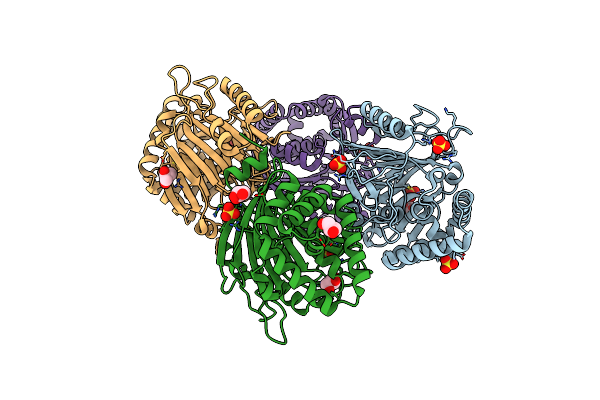
The Leua146Trp,Pheb24Tyr Double Mutant Of The Quorum Quenching N-Acyl Homoserine Lactone Acylase Pvdq Has An Altered Substrate Specificity Towards Small Acyl Chains
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: GOL |
 |
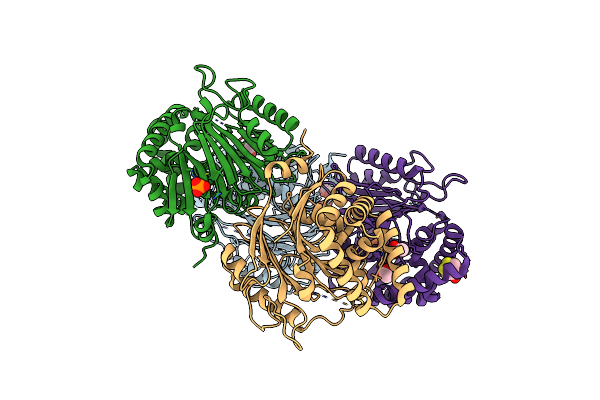
The Crystal Structure Of Precursor Acyl Coenzyme A:Isopenicillin N Acyltransferase From Penicillium Chrysogenum
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-03-09 Classification: TRANSFERASE Ligands: SO4, GOL, CL |
 |
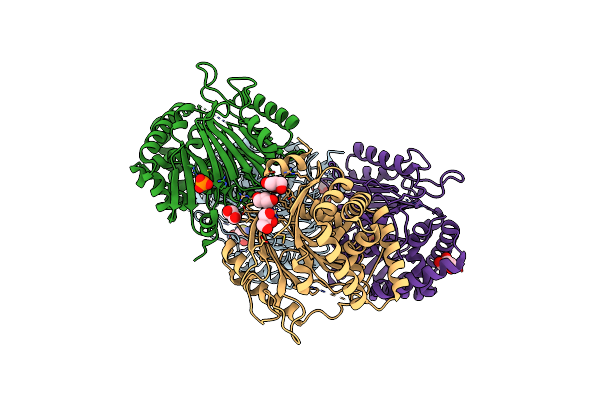
The Crystal Structure Of Mature Acyl Coenzyme A:Isopenicillin N Acyltransferase From Penicillium Chrysogenum
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2010-03-09 Classification: TRANSFERASE Ligands: PO4, GOL, CL, ACT, D1D |
 |
The Crystal Structure Of Mature Acyl Coenzyme A:Isopenicillin N Acyltransferase From Penicillium Chrysogenum In Complex 6- Aminopenicillanic Acid
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-03-09 Classification: TRANSFERASE Ligands: PO4, GOL, X1E, CL |

