Search Count: 14
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Acceptor Peptide And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Apo State, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Dol25-P-Man And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZY |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Ternary Complex With Dol25-P-C-Man And Acceptor Peptide, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZU |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: MEMBRANE PROTEIN Ligands: LMH |
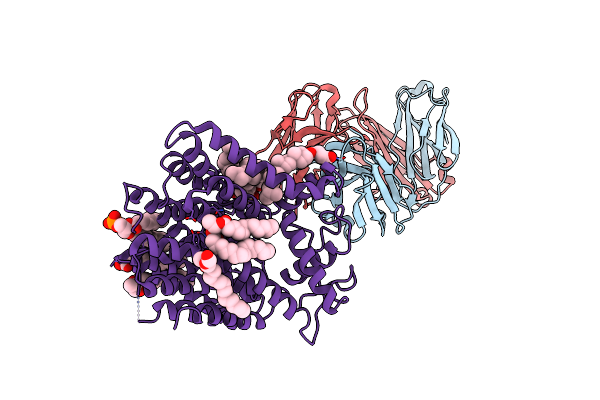 |
Cryo-Em Structure Of Nanodisc Reconstituted Yeast Alg6 In Complex With 6Ag9 Fab
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: MEMBRANE PROTEIN Ligands: PTY, Y01 |
 |
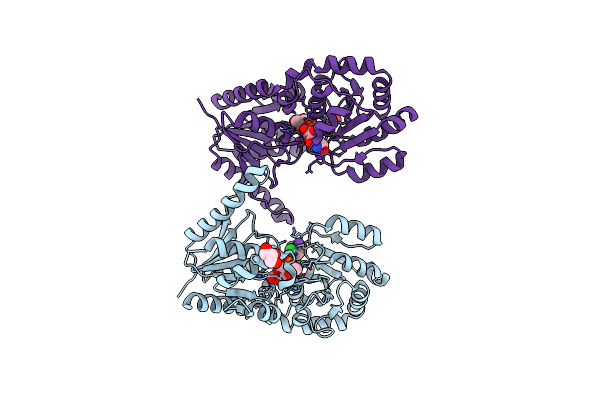
Bacterial Oligosaccharyltransferase Pglb In Complex With An Inhibitory Peptide And A Reactive Lipid-Linked Oligosaccharide Analog
Organism: Campylobacter lari (strain rm2100 / d67 / atcc baa-1060), Campylobacter lari rm2100
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN Ligands: MN, NA, MES, PEG, FFK |
 |
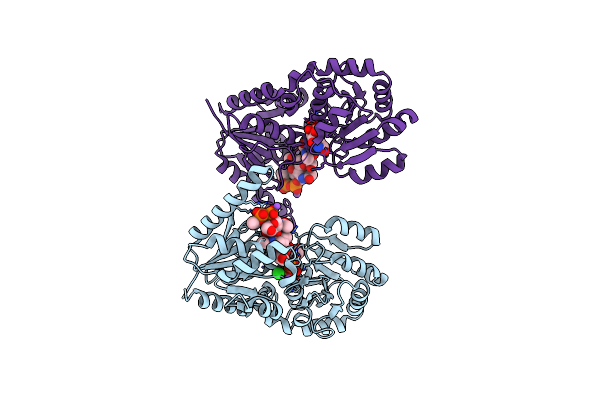
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: UD2, K, GOL, NA, CL |
 |
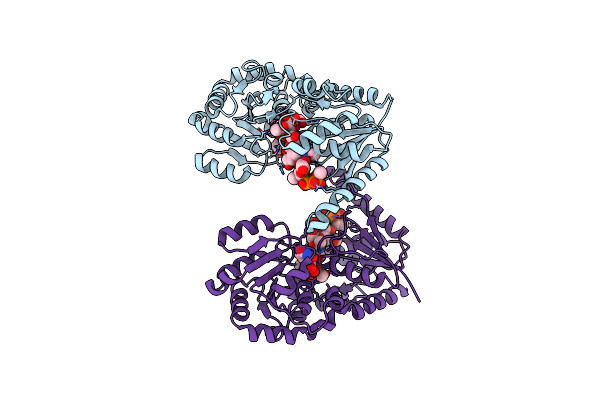
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: UDP, NA, CL, BUE |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: UDN, BUE |
 |
Structure Of Bacterial Oligosaccharyltransferase Pglb In Complex With An Acceptor Peptide And An Lipid-Linked Oligosaccharide Analog
Organism: Campylobacter lari (strain rm2100 / d67 / atcc baa-1060), Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-10-25 Classification: MEMBRANE PROTEIN Ligands: MN, 9UB, NA |
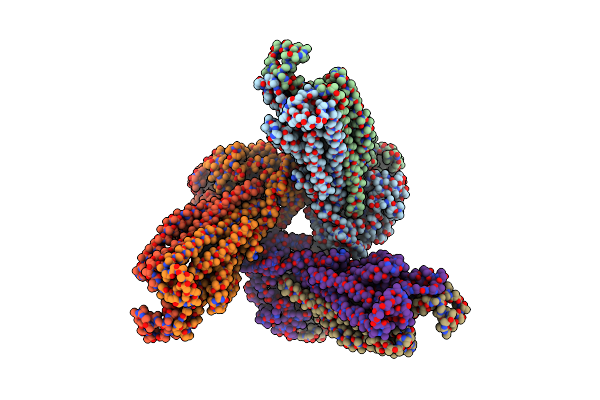 |
Atp-Driven Lipid-Linked Oligosaccharide Flippase Pglk In Outward-Occluded Conformation
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:5.90 Å Release Date: 2015-08-19 Classification: TRANSPORT PROTEIN |
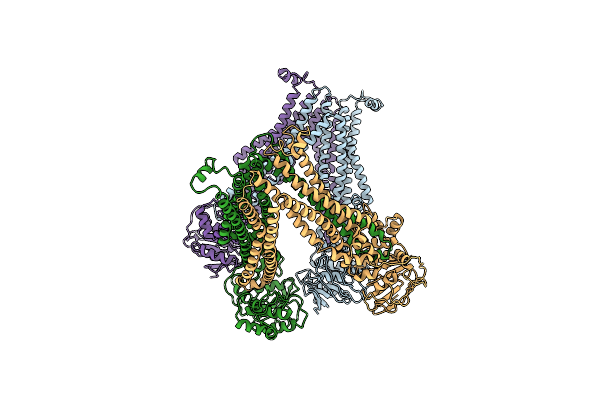 |
Atp-Driven Lipid-Linked Oligosaccharide Flippase Pglk In Apo-Inward Facing State (2)
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:3.94 Å Release Date: 2015-08-19 Classification: TRANSPORT PROTEIN |
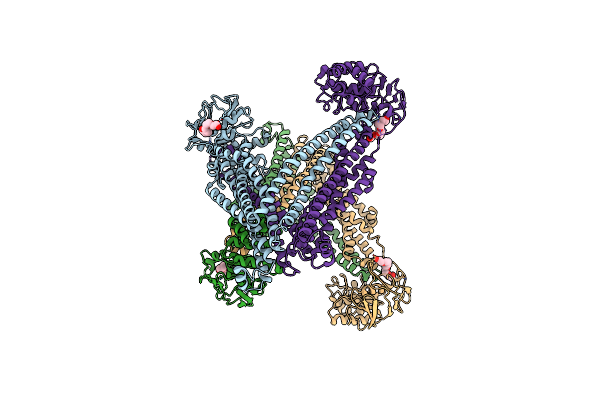 |
Atp-Driven Lipid-Linked Oligosaccharide Flippase Pglk In Apo-Inward State (1)
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-08-19 Classification: HYDROLASE Ligands: 1PE |

