Search Count: 39
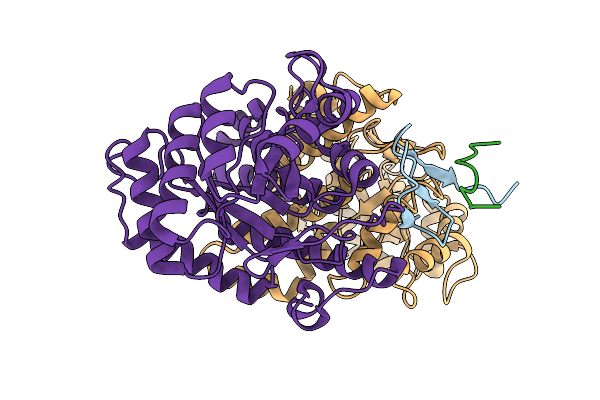 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1JAP, L1P, A1JAO, NA, A1JAN |
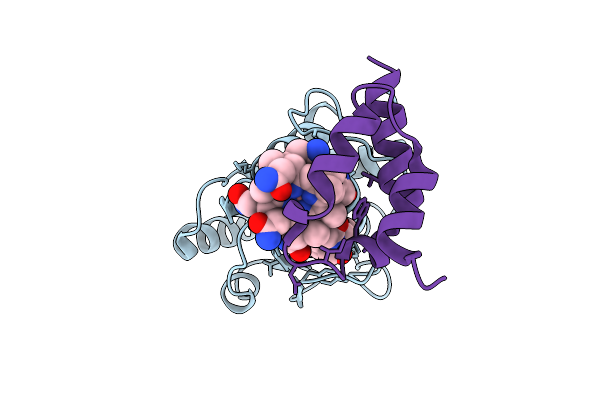 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: B13 |
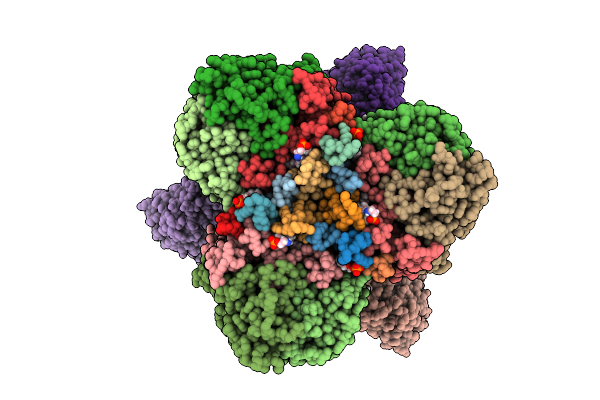 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: B13, A1JAP, L1P, A1JAO, NA, A1JAN |
 |
Cryo-Em Structure Of The Spike Glycoprotein From Bat Sars-Like Coronavirus (Bat Sl-Cov) Wiv1 In Locked State
Organism: Bat sars-like coronavirus wiv1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
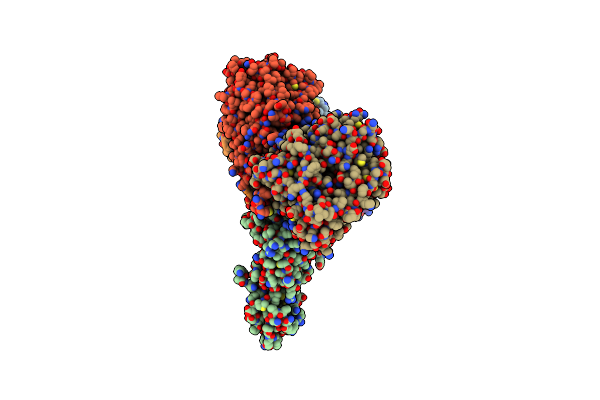 |
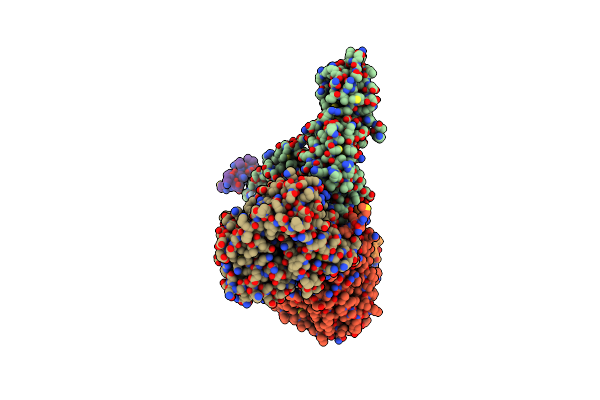
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Bound To Nadh
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: FES, SF4, FMN, NAD, RBF |
 |
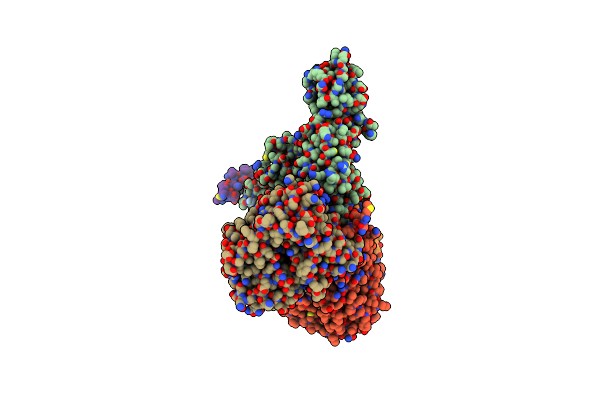
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Reduced With Low Potential Ferredoxin
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
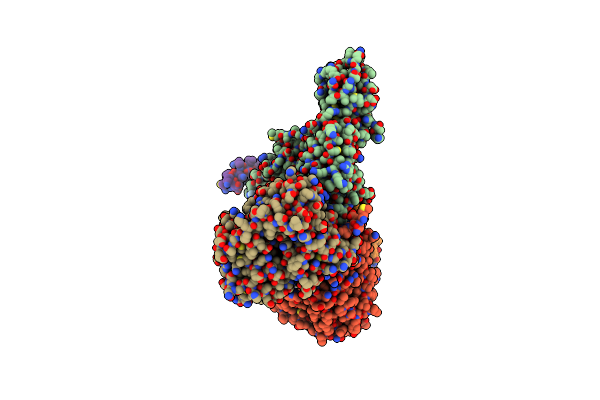
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Reduced With Low Potential Ferredoxin (Consensus Map)
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii In Apo State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
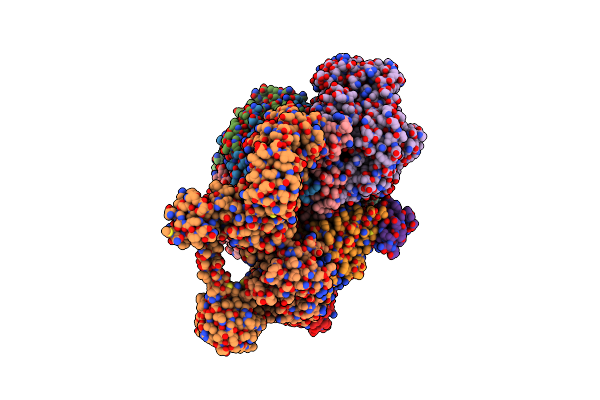 |
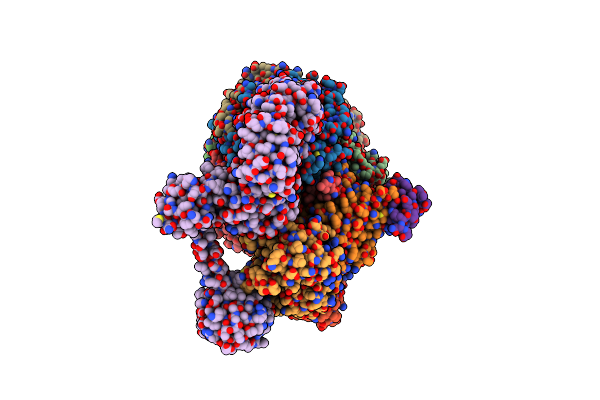
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
 |
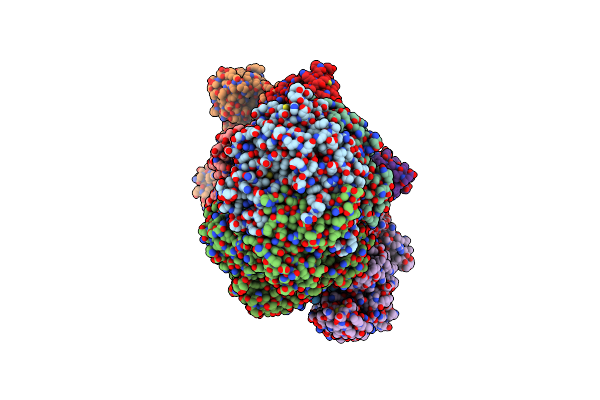
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
 |
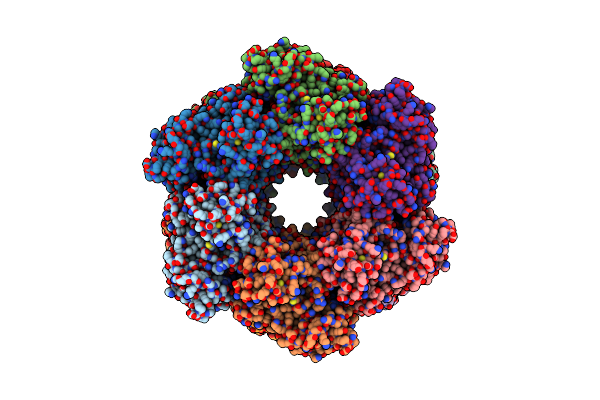
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
 |
Cryo-Em Structure Of The Active Dodecameric Methanosarcina Mazei Glutamine Synthetase.
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: CYTOSOLIC PROTEIN Ligands: AKG |
 |
Cryo-Em Structure Of Single-Layered Nucleoprotein-Rna Helical Assembly From Marburg Virus, Trimeric Repeat Unit
Organism: Marburg virus - musoke, kenya, 1980
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Methylophaga sulfidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-31 Classification: TRANSFERASE |
 |
Organism: Ananas comosus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE |
 |
Cryoem Structure Of A C7-Symmetrical Groel7-Groes7 Cage In Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Disordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Ordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Structure Average Of Groel14 Complexes Found In The Cytosol Of Escherichia Coli Overexpressing Groel Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |

