Search Count: 22
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Denuded Glycan Obtained By Soaking
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Nuded Glycan Obtained By Co-Crystallization
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Denuded Glycan Ended In Anhnam
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-13 Classification: CELL CYCLE Ligands: AH0 |
 |
1.65 Angstrom Resolution Crystal Structure Of Lmo0182 (Residues 1-245) From Listeria Monocytogenes Egd-E
Organism: Listeria monocytogenes serovar 1/2a (strain atcc baa-679 / egd-e)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: ACT |
 |
Cycloalternan-Forming Enzyme From Listeria Monocytogenes In Complex With Maltopentaose
Organism: Listeria monocytogenes serovar 1/2a (strain atcc baa-679 / egd-e)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-01-25 Classification: HYDROLASE Ligands: MG, CA, CL, PEG |
 |
Cycloalternan-Forming Enzyme From Listeria Monocytogenes In Complex With Panose
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-25 Classification: HYDROLASE Ligands: MG, CA, CL |
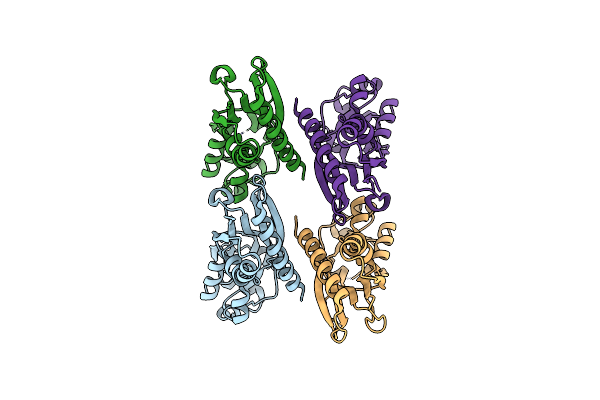 |
The X-Ray Structure Of The Effector Domain Of The Transcriptional Regulator Ampr Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-01-25 Classification: TRANSCRIPTION |
 |
Cycloalternan-Forming Enzyme From Listeria Monocytogenes In Complex With Cycloalternan
Organism: Listeria monocytogenes serovar 1/2a (strain atcc baa-679 / egd-e)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-12-14 Classification: SUGAR BINDING PROTEIN Ligands: MG, CA, CL, GLC |
 |
Cycloalternan-Degrading Enzyme From Trueperella Pyogenes In Complex With Isomaltose
Organism: Trueperella pyogenes
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-12-14 Classification: SUGAR BINDING PROTEIN Ligands: CA, MES |
 |
Cycloalternan-Degrading Enzyme From Trueperella Pyogenes In Complex With Covalent Intermediate
Organism: Trueperella pyogenes
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-12-14 Classification: SUGAR BINDING PROTEIN Ligands: CA, PGE |
 |
Cycloalternan-Degrading Enzyme From Trueperella Pyogenes In Complex With Cycloalternan
Organism: Trueperella pyogenes
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-12-14 Classification: SUGAR BINDING PROTEIN Ligands: SIN |
 |
Structure Of Lytic Transglycosylase Mltc From Escherichia Coli At 2.3 A Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-07-23 Classification: LYASE |
 |
Structure Of Lytic Transglycosylase Mltc From Escherichia Coli In Complex With Tetrasaccharide At 2.9 A Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-07-23 Classification: HYDROLASE |
 |
Crystal Structure Of Mltc In Complex With Tetrasaccharide At 2.15 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-07-23 Classification: LYASE Ligands: P6G, CIT |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-07-23 Classification: LYASE Ligands: NAG, AH0 |
 |
Crystal Structure Of Ampdh3 From Pseudomonas Aeruginosa In Complex With Tetrasaccharide Pentapeptide
Organism: Pseudomonas aeruginosa pao1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-10-09 Classification: HYDROLASE/PEPTIDE Ligands: ZN |
 |
Crystal Structure Of Ampdh3 From Pseudomonas Aeruginosa In Complex With Anhydromuramic Pentapeptide
Organism: Pseudomonas aeruginosa pao1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-10-09 Classification: HYDROLASE/PEPTIDE Ligands: ACT, PEG |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: FLC |

