Search Count: 40
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2023-06-21 Classification: ANTIBIOTIC Ligands: MOH |
 |
Bacillus Cereus Dna Glycosylase Alkd Bound To A Cc1065-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNG |
 |
Bacillus Cereus Dna Glycosylase Alkd Bound To A Duocarmycin Sa-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNY |
 |
Organism: Homo sapiens, Eptatretus stoutii
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-04-17 Classification: IMMUNE SYSTEM/AGONIST Ligands: KQD, NAG |
 |
Organism: Homo sapiens, Eptatretus stoutii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-17 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
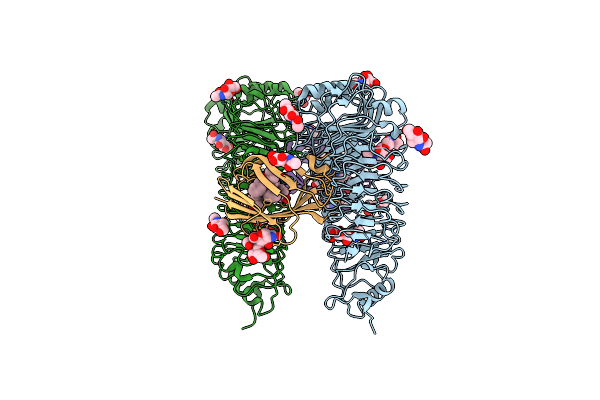
Organism: Mus musculus, Eptatretus burgeri
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2016-04-27 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Mus musculus, Eptatretus burgeri
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2016-04-27 Classification: IMMUNE SYSTEM Ligands: NAG, V2A |
 |
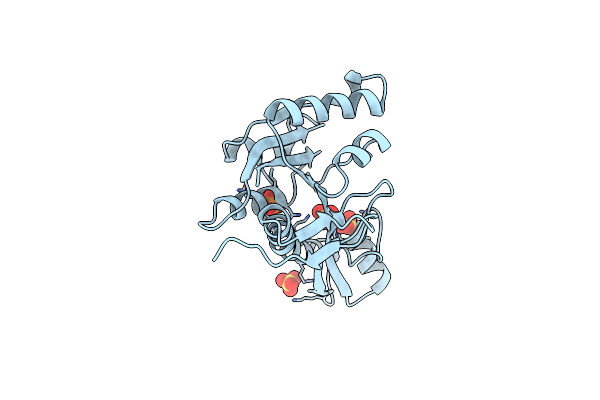
Organism: Mus musculus, Eptatretus burgeri
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-04-27 Classification: IMMUNE SYSTEM Ligands: NAG, LP4, LP5, DAO, MYR |
 |
High Resolution Structure Of Human Glycinamide Ribonucleotide Transformylase In Apo Form.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2013-07-31 Classification: TRANSFERASE Ligands: PO4, SO4 |
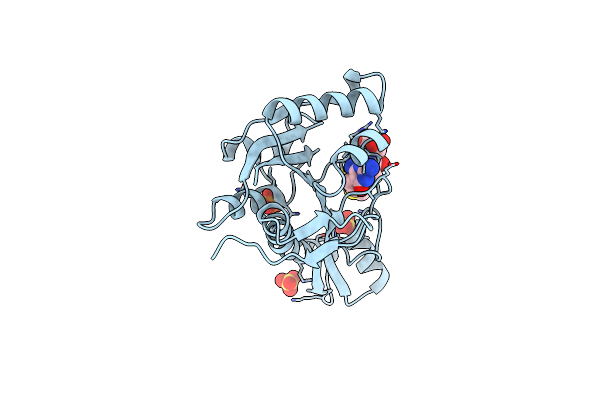 |
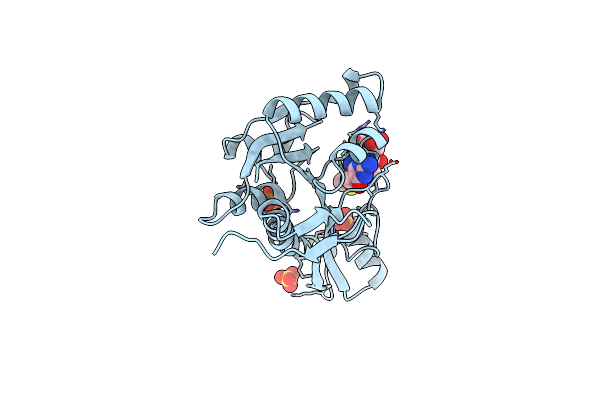
The Structure Of Human Glycinamide Ribonucleotide Transformylase In Complex With 10S-Methylthio-Ddathf.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-07-31 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: PO4, SO4, DXY |
 |
The Structure Of Human Glycinamide Ribonucleotide Transformylase In Complex With 10R-Methylthio-Ddathf.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-31 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: PO4, SO4, DXZ |
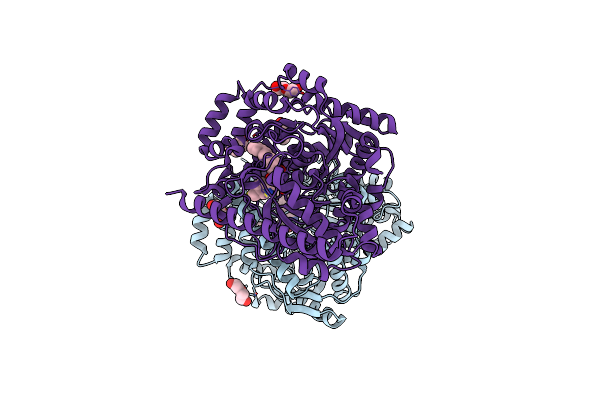 |
Crystal Structure Of A Covalently Bound Alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) To A Humanized Variant Of Fatty Acid Amide Hydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BR1, PEG, CL |
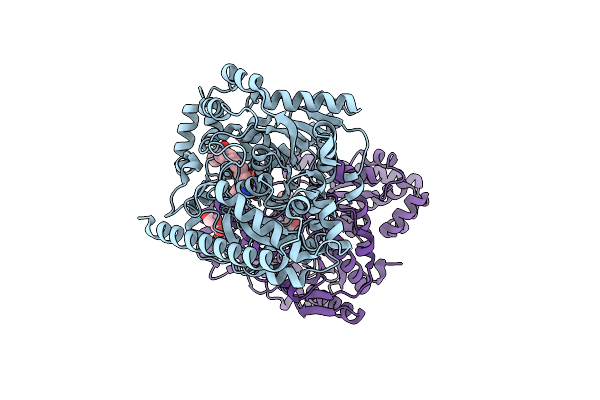 |
Crystal Structure Of A Covalently Bound Alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) To A Humanized Variant Of Fatty Acid Amide Hydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-11-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: JG2, CL, PEG |
 |
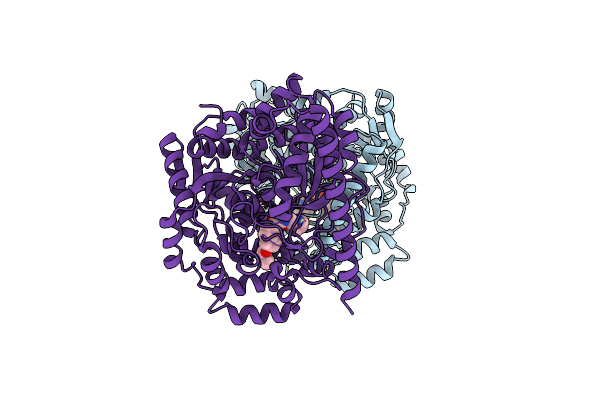
Crystal Structure Of A Noncovalently Bound Alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) To A Humanized Variant Of Fatty Acid Amide Hydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2011-11-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: JG1, CL, F, PEG, 1DO |
 |
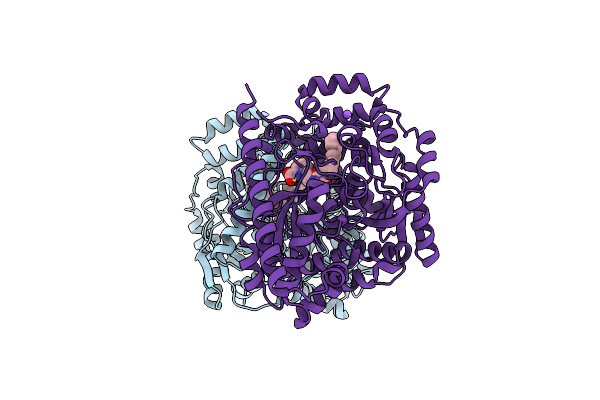
Alpha-Ketoheterocycle Inhibitors Of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints In The Acyl Side Chain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-06 Classification: Hydrolase/Hydrolase Inhibitor Ligands: OJ8, CL |
 |
Crystal Structure Analysis Of A Phenhexyl/Oxazole/Carboxypyridine Alpha-Ketoheterocycle Inhibitor Bound To A Humanized Variant Of Fatty Acid Amide Hydrolase'
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-12-01 Classification: HYDROLASE Ligands: F2C, CL, NA |
 |
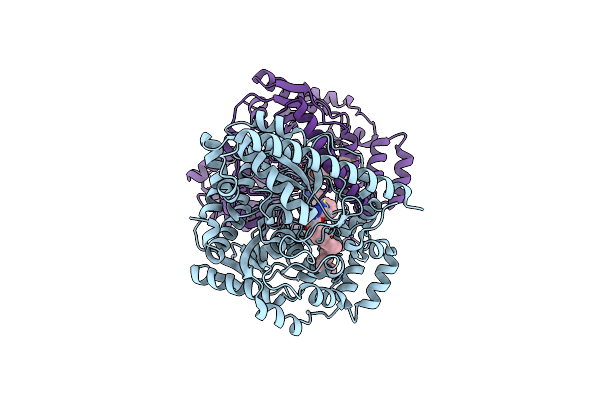
Crystal Structure Analysis Of A Biphenyl/Oxazole/Carboxypyridine Alpha-Ketoheterocycle Inhibitor Bound To A Humanized Variant Of Fatty Acid Amide Hydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-12-01 Classification: HYDROLASE Ligands: F27, CL, 1DO |
 |
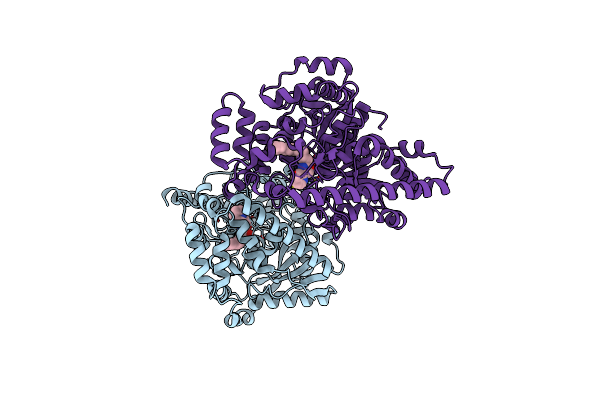
Crystal Structure Analysis Of A Oleyl/Oxadiazole/Pyridine Inhibitor Bound To A Humanized Variant Of Fatty Acid Amide Hydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-12-01 Classification: HYDROLASE Ligands: K84, CL |
 |
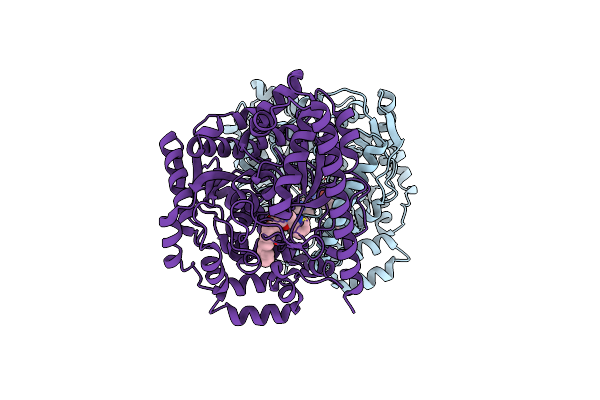
3D-Crystal Structure Of Humanized-Rat Fatty Acid Amide Hydrolase (Faah) Conjugated With 7-Phenyl-1-(4-(Pyridin-2-Yl)Oxazol-2-Yl)Heptan- 1-One, An Alpha-Ketooxazole
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2009-09-15 Classification: HYDROLASE Ligands: S99, CL |
 |
3D-Crystal Structure Of Humanized-Rat Fatty Acid Amide Hydrolase (Faah) Conjugated With 7-Phenyl-1-(5-(Pyridin-2-Yl)Oxazol-2-Yl)Heptan- 1-One, An Alpha-Ketooxazole
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2009-09-15 Classification: HYDROLASE Ligands: OL1, CL |

