Search Count: 24
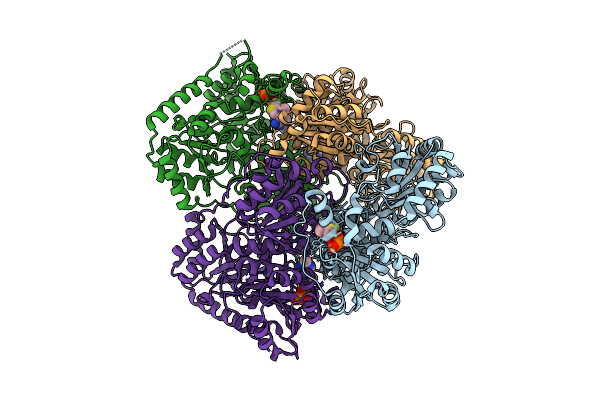 |
Ancestrally Reconstructed Acetolactate Synthase - Ancestor N259 With Bound Thdp And Magnesium.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: TPP, MG |
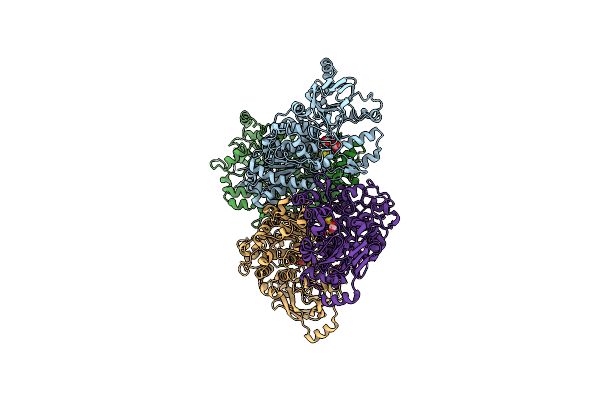 |
Crystal Structure Of Fe-S Cluster-Dependent Dehydratase From Paralcaligenes Ureilyticus In Complex With Mg
Organism: Paralcaligenes ureilyticus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-08-30 Classification: LYASE Ligands: FES, MG, BCT, CO2 |
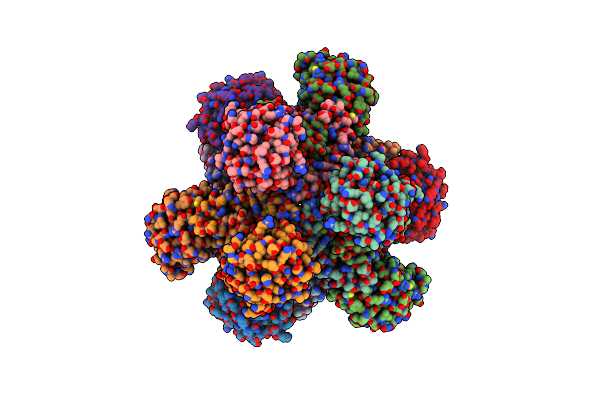 |
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
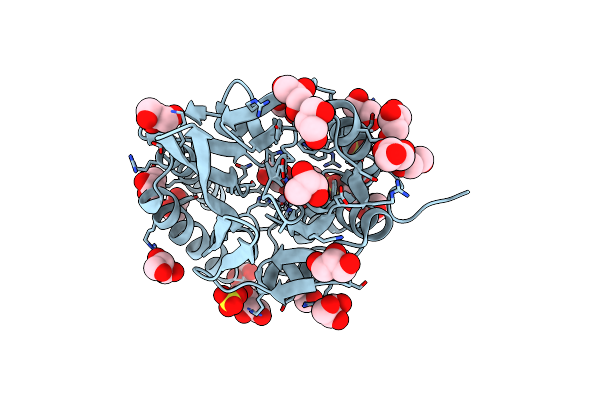 |
Kinetic And Structural Characterization Of The First B3 Metallo-Beta-Lactamase With An Active Site Glutamic Acid
Organism: Sphingobium indicum (strain dsm 16412 / ccm 7286 / mtcc 6364 / b90a)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-07-28 Classification: HYDROLASE Ligands: ZN, SO4, GOL |
 |
Crystal Structure Of The Tir Domain From Human Sarm1 In Complex With Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: BDR, CL |
 |
Crystal Structure Of The Tir Domain From Human Sarm1 In Complex With Glycerol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Selenomethionine Labelled Tandem Sam Domains (L446M:L505M:L523M Mutant) From Human Sarm1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Tir Domain G601P Mutant From Human Sarm1, Crystal Form 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: CL, MES |
 |
Crystal Structure Of The Tir Domain From The Grapevine Disease Resistance Protein Run1 In Complex With Nadp+ And Bis-Tris
Organism: Vitis rotundifolia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: A2P, BTB |
 |
Crystal Structure Of The Tir Domain G601P Mutant From Human Sarm1, Crystal Form 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: MES |
 |
Crystal Structure Of The Tir Domain From The Muscadinia Rotundifolia Disease Resistance Protein Rpv1
Organism: Vitis rotundifolia
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: MLA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-08-21 Classification: TRANSPORT PROTEIN/HYDROLASE |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-08-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-08-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-08-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-09 Classification: TRANSPORT PROTEIN |
 |
Organism: Oryza sativa, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-01-09 Classification: TRANSPORT PROTEIN |

