Search Count: 121
 |
X-Ray Crystal Structure Of Streptomyces Cacaoi Pold With Iron And Succinate Bound
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FE, SO4, SIN |
 |
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: ZN, GOL |
 |
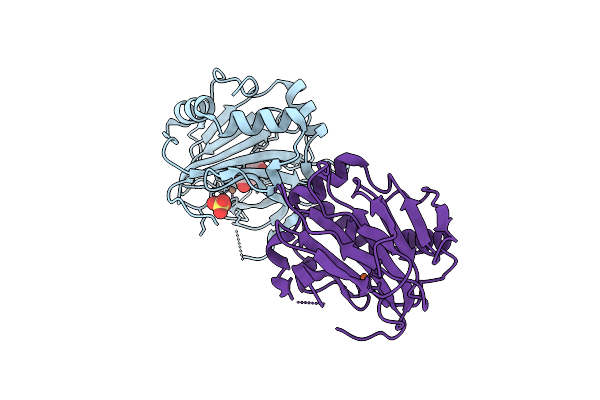
X-Ray Crystal Structure Of Streptomyces Cacaoi Polf In Complex With Iron And L-Isoleucine
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: GOL, FE2, ZN, ILE |
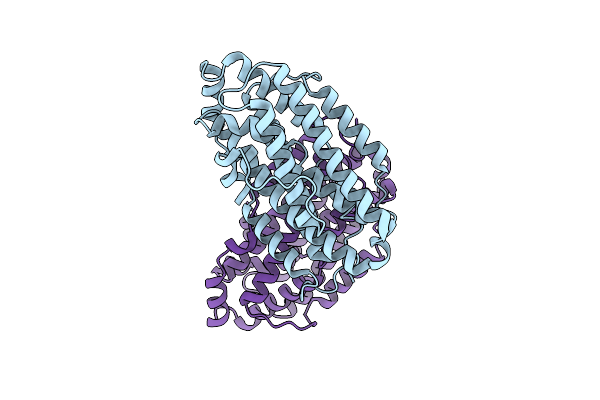 |
X-Ray Crystal Structure Of Francisella Hispaniensis Apo Ribonucleotide Reductase Beta Subunit
Organism: Francisella hispaniensis
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: CL |
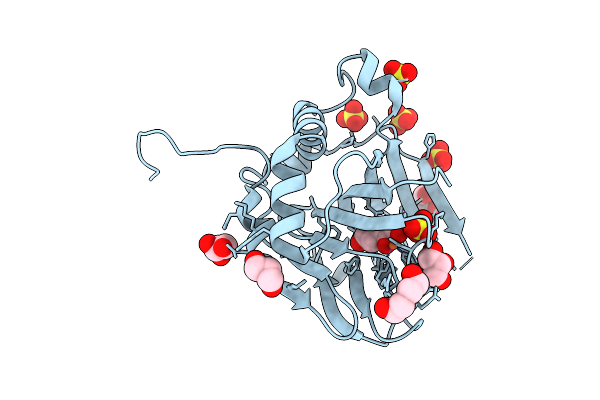 |
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: AKG, SO4, GOL, PEG, FE |
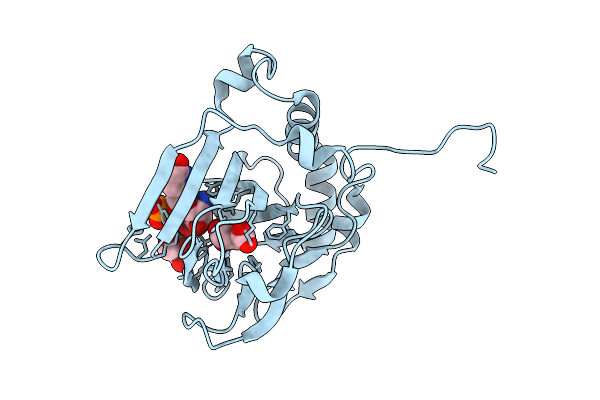 |
Pasi From Photorhabdus Asymbiotica Bound To Vanadyl, Succinate, And 5-Amino-6-Hydroxy-Octanosyl Acid 2-Phosphate
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: A1BIK, GOL, SIN, CL, V |
 |
Organism: Methylorubrum extorquens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-16 Classification: METAL BINDING PROTEIN |
 |
Organism: Methylorubrum extorquens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-16 Classification: METAL BINDING PROTEIN Ligands: LA |
 |
Organism: Methylorubrum extorquens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-16 Classification: METAL BINDING PROTEIN Ligands: CE |
 |
Organism: Methylorubrum extorquens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-10-16 Classification: METAL BINDING PROTEIN Ligands: EU |
 |
Organism: Methylorubrum extorquens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-10-16 Classification: METAL BINDING PROTEIN Ligands: HO3 |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SR, EDO, FMT, FE2, AKG, HYO |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, HYO, SIN, FMT, V, O, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OVR, SIN, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, AKG, OVR, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, SIN, FMT, OVR, V, O, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And Scopolamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OW0, SIN, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, HYO, AKG, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, FMT, HYO, SIN, V, O, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, AKG, OVR, SR |

