Search Count: 69
 |
Organism: Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-10-29 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-10-29 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-10-29 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-10-29 Classification: IMMUNE SYSTEM Ligands: EDO, IPA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-29 Classification: IMMUNE SYSTEM Ligands: EDO, PEG |
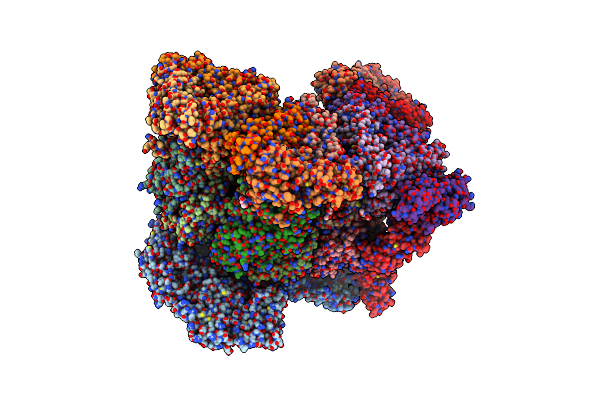 |
Organism: Rotavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRUS Ligands: CA, NAG, ZN |
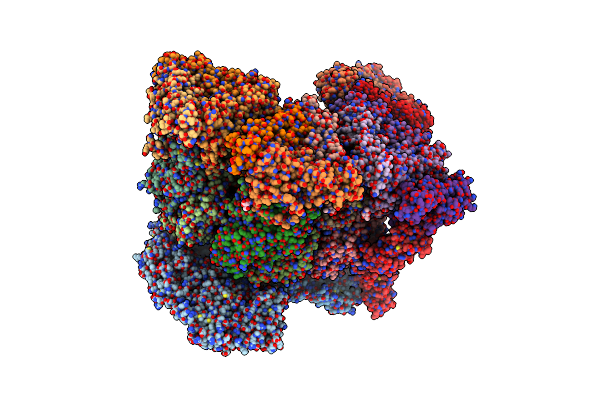 |
Organism: Rotavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRUS Ligands: CA, NAG, ZN |
 |
Cryo-Em Reconstruction Of Vp4 Assembly From Sa11 Rotavirus Non-Tripsinized Triple Layered Particle
|
 |
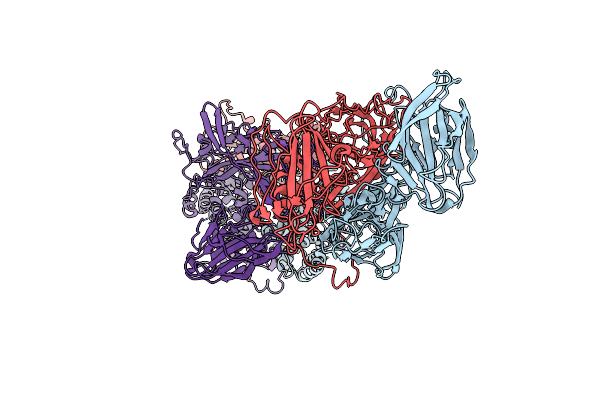
Cryo-Em Reconstruction Of Vp5*/Vp8* Assembly From Sa11 Rotavirus Tripsinized Triple Layered Particle
|
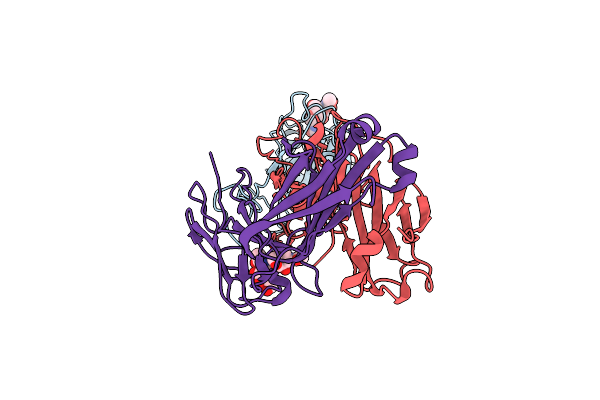 |
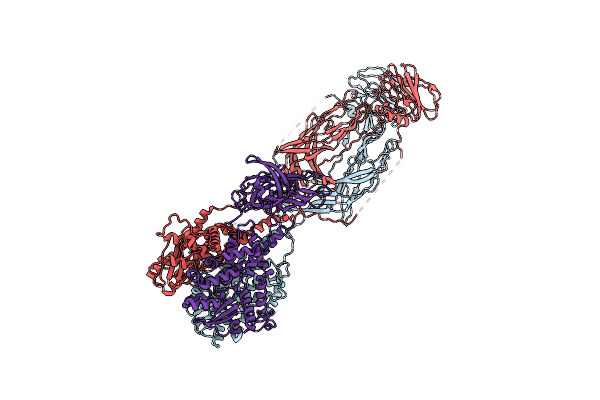
Sars-Cov-2 Spike In Complex With The 17T2 Neutralizing Antibody Fab Fragment (Local Refinement Of Rbd And Fab)
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: VIRAL PROTEIN |
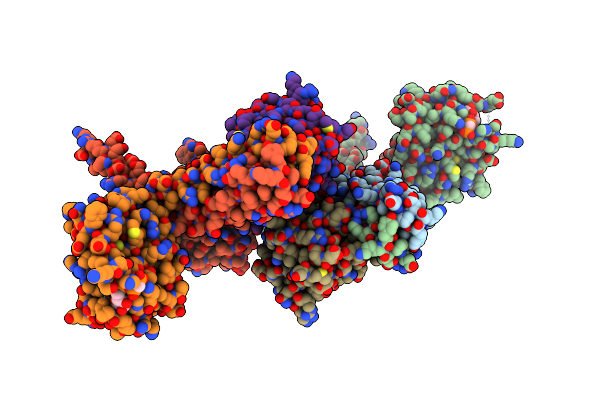 |
Crystal Structure Of The Desk:Desr-Q10A Complex In The Phosphotransfer State
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: ACP, MG |
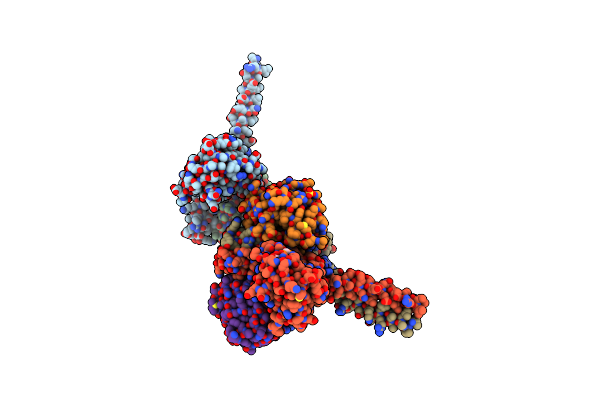 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: ACP, MG, BEF, SO4 |
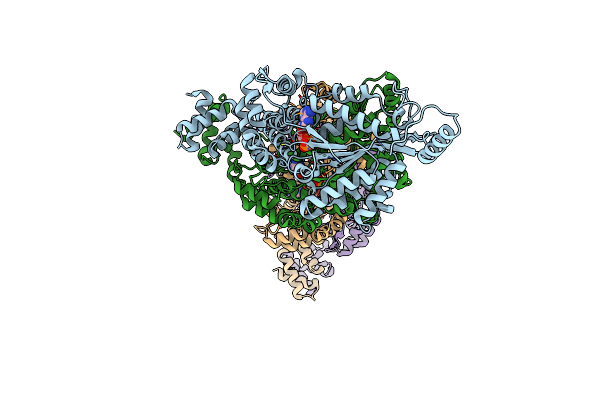 |
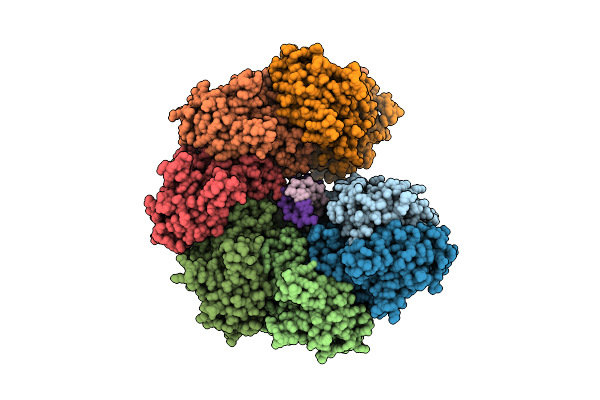
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-02-23 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
 |
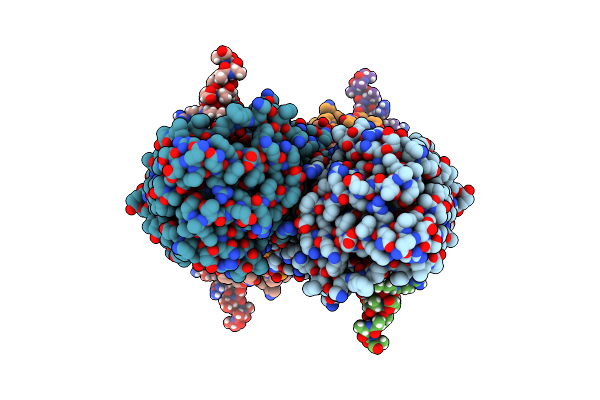
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-02-23 Classification: DNA BINDING PROTEIN/DNA Ligands: ANP, MG |
 |
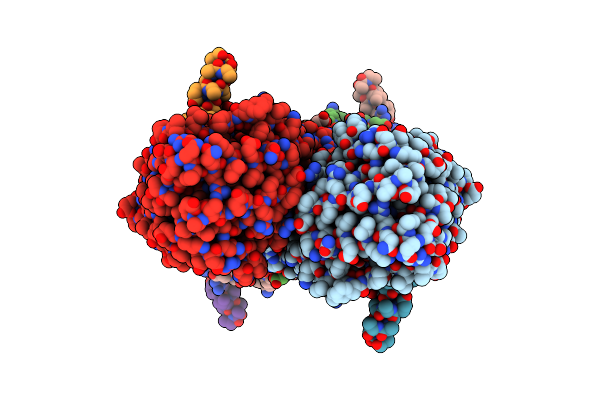
Partial Structure Of Tyrosine Hydroxylase Lacking The First 35 Residues In Complex With Dopamine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
 |
Partial Structure Of Tyrosine Hydroxylase In Complex With Dopamine Showing The Catalytic Domain And An Alpha-Helix From The Regulatory Domain Involved In Dopamine Binding.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
 |
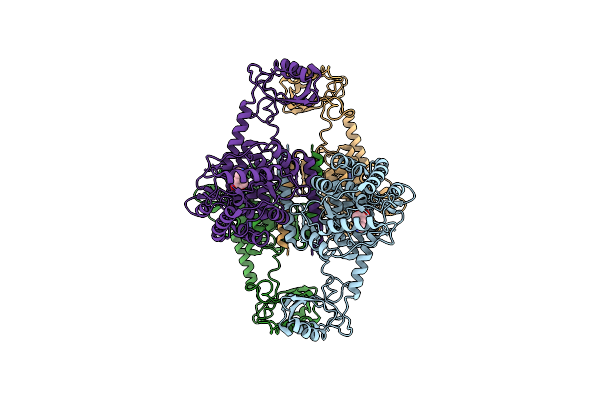
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Atomic Model Of The Em-Based Structure Of The Full-Length Tyrosine Hydroxylase In Complex With Dopamine (Residues 40-497) In Which The Regulatory Domain (Residues 40-165) Has Been Included Only With The Backbone Atoms
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE, LDP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-10-21 Classification: RIBOSOME Ligands: MG |

