Search Count: 13
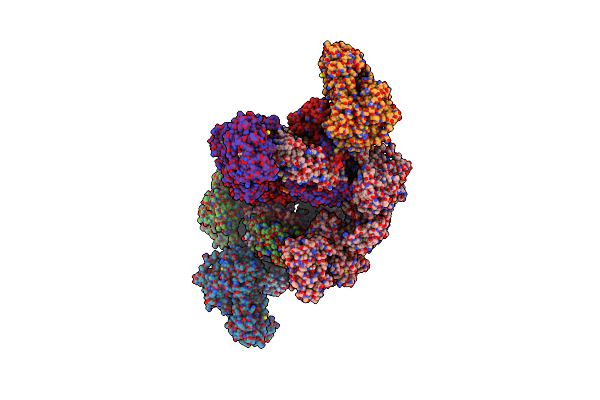 |
Structure Of The Human Atp Citrate Lyase Holoenzyme In Complex With Citrate, Coenzyme A And Mg.Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-04-10 Classification: LYASE Ligands: MG, PO4, FLC, COA, ADP |
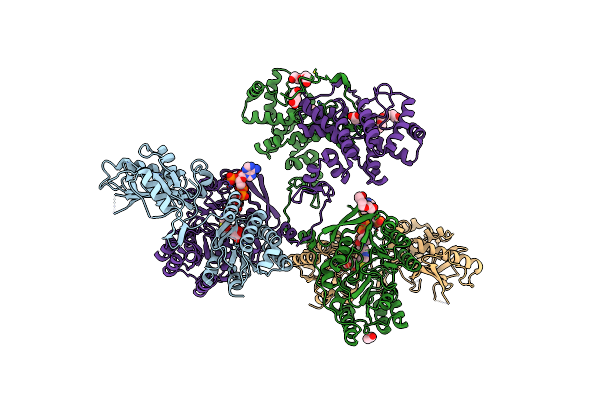 |
Structure Of Atp Citrate Lyase From Methanothrix Soehngenii In Complex With Citrate And Coenzyme A
Organism: Methanosaeta concilii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, NA, COA, PGE, PG4, ACT, SIN |
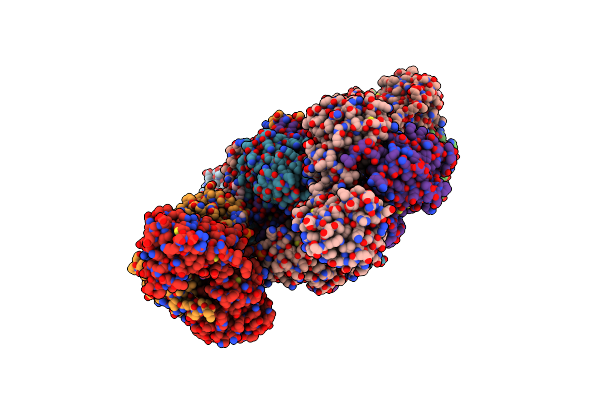 |
Structure Of Atp Citrate Lyase From Chlorobium Limicola In Complex With Citrate And Coenzyme A.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, COA, PGE, TRS, PG4 |
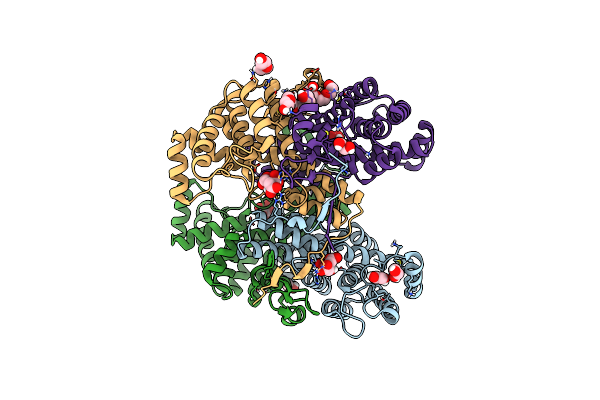 |
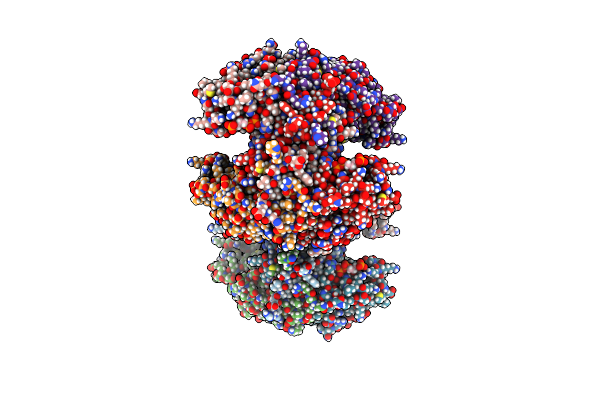
Structure Of The Citryl-Coa Lyase Core Module Of Human Atp Citrate Lyase In Complex With Citrate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, EDO, PGE, 1PE, PEG |
 |
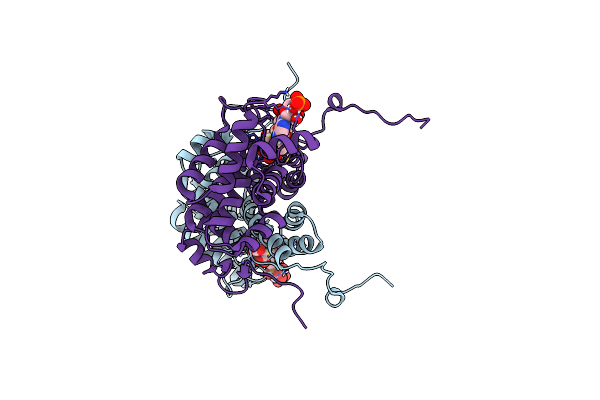
Structure Of The Citryl-Coa Lyase Core Module Of Human Atp Citrate Lyase In Complex With Citrate And Coash (Space Group P21)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-04-10 Classification: LYASE Ligands: COA, FLC, PEG, SO4, CAO, EDO |
 |
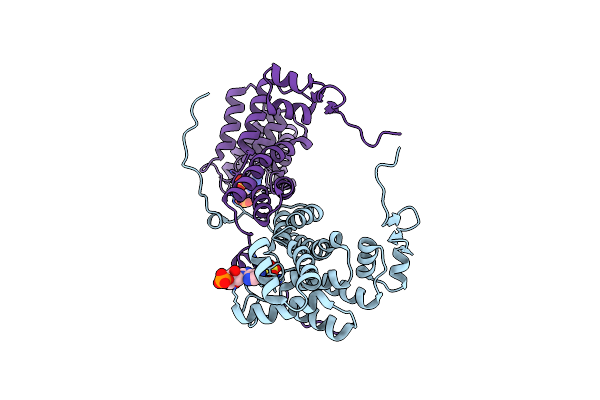
Structure Of The Citryl-Coa Lyase Core Module Of Human Atp Citrate Lyase In Complex With Citrate And Coash In Space Group C2221
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-04-10 Classification: LYASE Ligands: COA, FLC |
 |
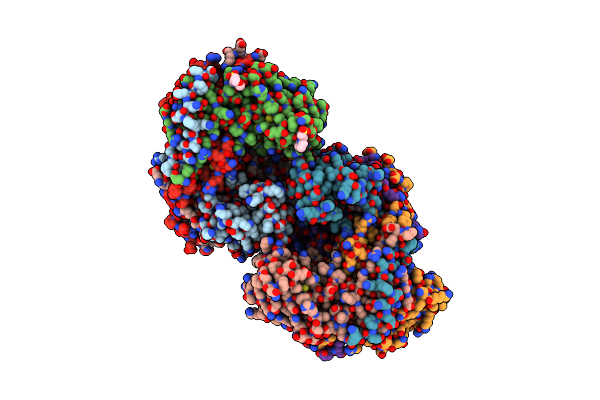
Structure Of The Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase (Space Group P3121)
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-04-10 Classification: LYASE Ligands: COA, SO4 |
 |
Structure Of The Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase (Space Group P21)
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, MES |
 |
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-04-10 Classification: LYASE Ligands: COA, SO4 |
 |
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2019-04-10 Classification: LYASE Ligands: PGE, COA, FLC |
 |
Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase In Complex With Acetyl-Coa And L-Malate
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-04-10 Classification: LYASE Ligands: CA, LMR, ACO, PG4, PGE |
 |
Structure Of The Human Atp Citrate Lyase Holoenzyme In Complex With Citrate, Coenzyme A And Mg.Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2019-04-10 Classification: LYASE Ligands: 2HP, COA, ADP, MG, FLC |
 |
Organism: Bacillus subtilis
Method: SOLID-STATE NMR Release Date: 2008-06-17 Classification: TRANSPORT PROTEIN |

