Search Count: 152
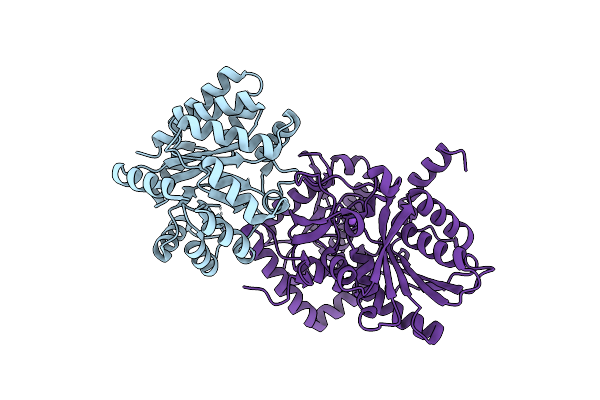 |
Joint X-Ray/Neutron Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine From Microgravity-Grown Crystal
Organism: Salmonella enterica subsp. enterica serovar typhi, Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.80 Å, 2.10 Å Release Date: 2024-02-14 Classification: LYASE |
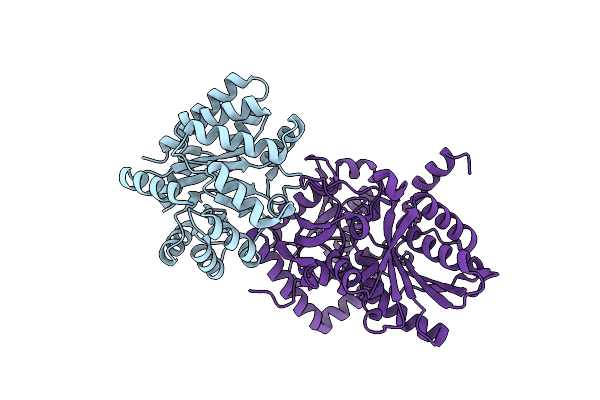 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine At Ph 5.0
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE |
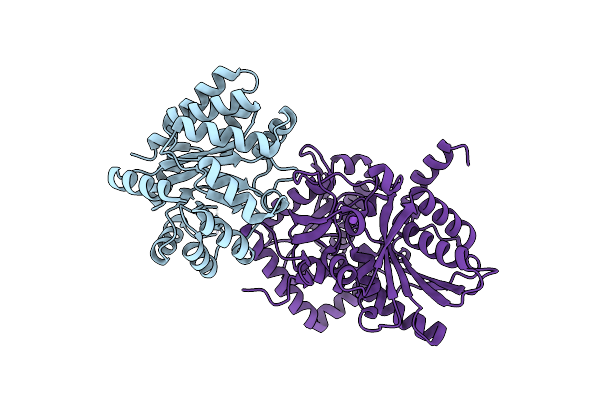 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-14 Classification: LYASE |
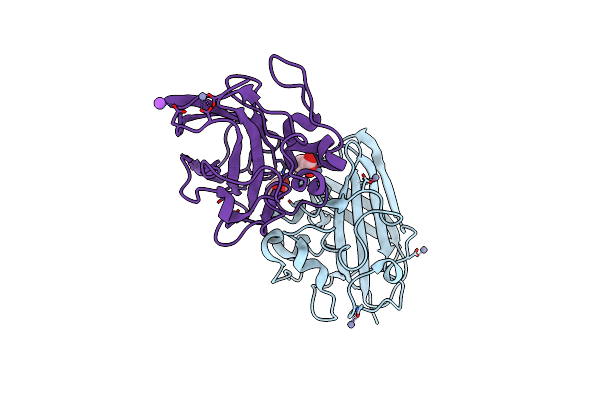 |
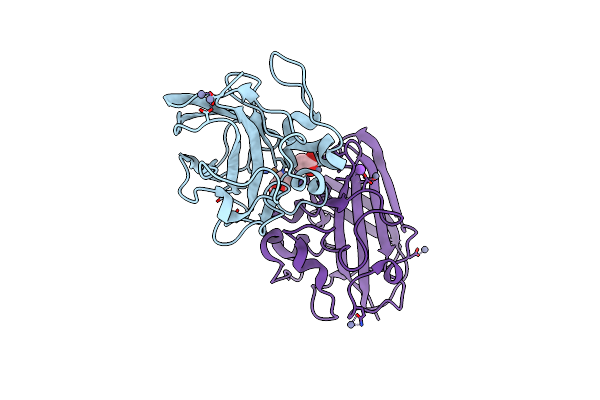
Organism: Vibrio cholerae o1 biovar el tor
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2023-08-30 Classification: CELL ADHESION Ligands: ACT, CU, ZN, NA |
 |
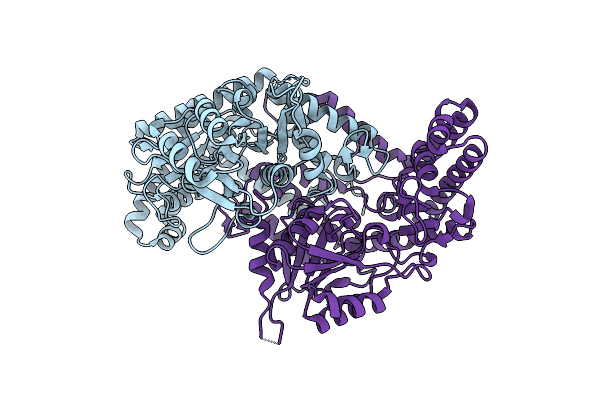
Organism: Vibrio cholerae o1 biovar el tor
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-08-30 Classification: CELL ADHESION Ligands: ACT, CU, ZN, NA |
 |
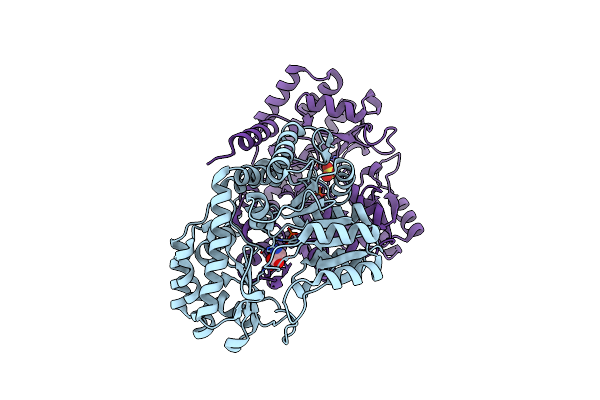
Room-Temperature X-Ray Structure Of Human Mitochondrial Serine Hydroxymethyltransferase (Hshmt2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: CL |
 |
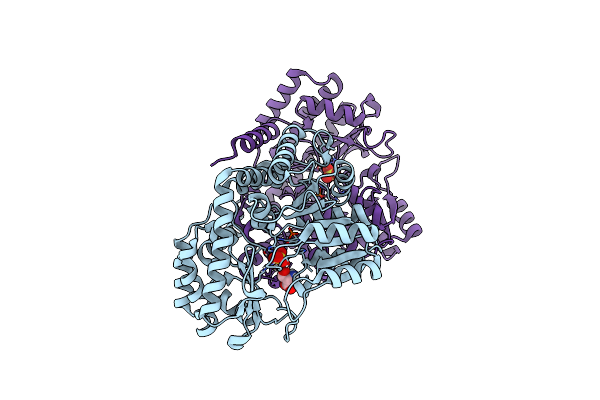
Room-Temperature X-Ray Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Shmt) Bound With D-Ser In A Pseudo-Michaelis Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: DSN, SO4 |
 |
Joint X-Ray/Neutron Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Tthshmt) In Internal Aldimine State With L-Ser Bound In A Pre-Michalis Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.30 Å, 2.00 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, SER, DOD |
 |
Joint X-Ray/Neutron Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Tthshmt) In Internal Aldimine State
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.30 Å, 2.00 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, DOD |
 |
Joint X-Ray/Neutron Structure Of Aspastate Aminotransferase (Aat) In Complex With Pyridoxamine 5'-Phosphate (Pmp)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.70 Å, 2.22 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: PLA, PMP, DOD |
 |
Joint X-Ray/Neutron Structure Of Sars-Cov-2 Main Protease (3Cl Mpro) In Complex With Bbh-1
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.85 Å, 2.20 Å Release Date: 2022-03-02 Classification: HYDROLASE/INHIBITOR Ligands: I1W, DOD |
 |
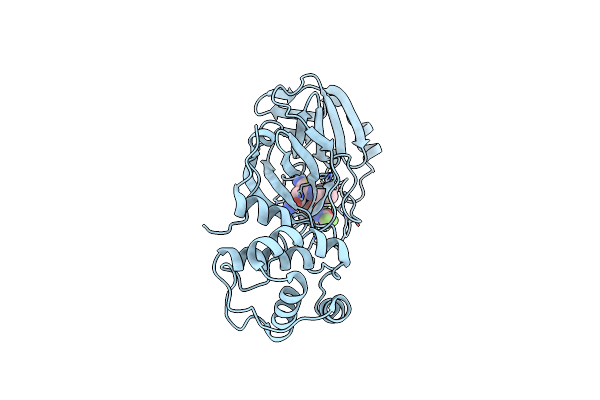
Room Temperature X-Ray Structure Of Sars-Cov-2 Main Protease (3Cl Mpro) In Complex With Bbh-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-02 Classification: HYDROLASE/INHIBITOR Ligands: I1Z |
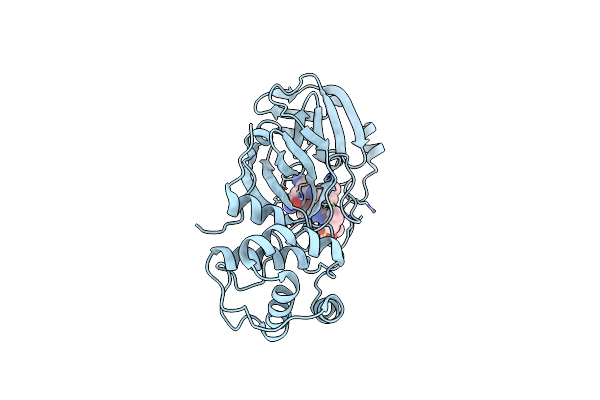 |
Room Temperature X-Ray Structure Of Sars-Cov-2 Main Protease (3Cl Mpro) In Complex With Nbh-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-02 Classification: HYDROLASE/INHIBITOR Ligands: NB2 |
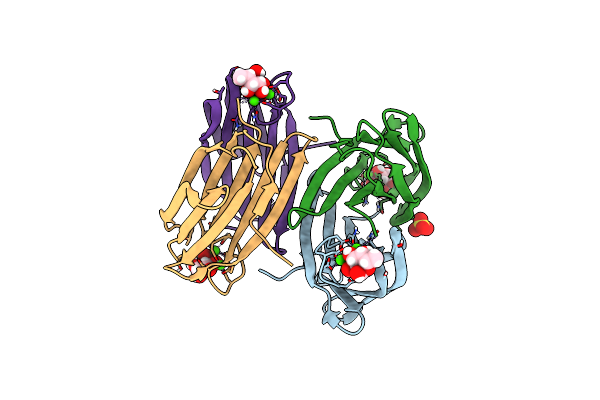 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Lecb Lectin In Complex With Perdeuterated Fucose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.85 Å, 1.90 Å Release Date: 2022-01-12 Classification: SUGAR BINDING PROTEIN Ligands: CA, FUC, SO4 |
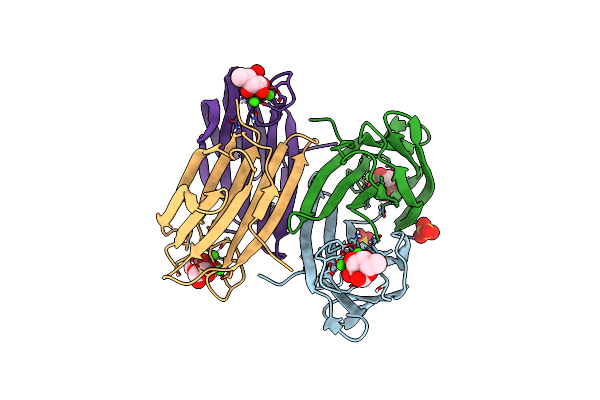 |
X-Ray Crystal Structure Of Perdeuterated Lecb Lectin In Complex With Perdeuterated Fucose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2022-01-12 Classification: SUGAR BINDING PROTEIN Ligands: FUC, CA, SO4 |
 |
Room Temperature X-Ray Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With Pf-07321332
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: 4WI |
 |
Room Temperature Structure Of Hache In Complex With Substrate Analog 4K-Tma
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-09-22 Classification: HYDROLASE Ligands: NWA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-22 Classification: HYDROLASE Ligands: NWA, GOL, NO3 |
 |
Room Temperature Structure Of Hache In Complex With Substrate Analog 4K-Tma And Mmb4 Oxime
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-09-22 Classification: HYDROLASE Ligands: NWA, 3VI |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |

