Search Count: 18
 |
Human Fatty Acid Binding Protein 4 In Complex With 6-Chloro-2-Methyl-4-Phenyl-Quinoline-3-Carboxylic Acid At 1.18A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2016-03-09 Classification: LIPID BINDING PROTEIN Ligands: 5M8 |
 |
Human Fabp4 In Complex With 6-Chloro-4-Phenyl-2-Piperidin-1-Yl-Quinoline-3-Carboxylic Acid At 1.29A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-03-09 Classification: LIPID BINDING PROTEIN Ligands: 5M7, SO4 |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C13C(Cc(S1)C(Ncc2Occc2)=O)C(Nn3C4Ccc(Cc4)Cl)C, Micromolar Ic50=0.217
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, GOL, 5M6 |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C1(C(Nc([Nh]1)Cl)C2Ccccc2)C4=Nn(C3Cccc(C3)Oc(F)(F)F)C=Cc4=O, Micromolar Ic50=0.029618
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5MG |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With N4C(C)N1C(Nc(N1)Ccc2Nc(Nn2C)N3Cccc3)C(C4)Cc, Micromolar Ic50=0.0037753
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5MF |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C2(C(N1Nc(Nc1C(C2)C)Ccc3Nc(Nn3C)N4Cccc4)C)Cl, Micromolar Ic50=0.000279
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5M9 |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C2(=Nn(C1Cccc(C1)Oc(F)(F)F)C=Cc2=O)C3Ccnn3C4Ccccc4, Micromolar Ic50=0.019462
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 67A |
 |
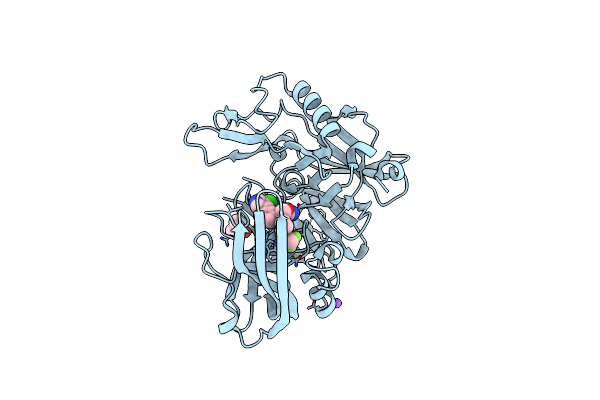
Crystal Structure Of Bace-1 In Complex With {(1R,2R)-2-[(R)-2-Amino-4-(4-Difluoromethoxy-Phenyl)-4,5-Dihydro-Oxazol-4-Yl]-Cyclopropyl}-(5-Chloro-Pyridin-3-Yl)-Methanone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-24 Classification: Hydrolase/Inhibitor Ligands: 5T5, NA, DMS |
 |
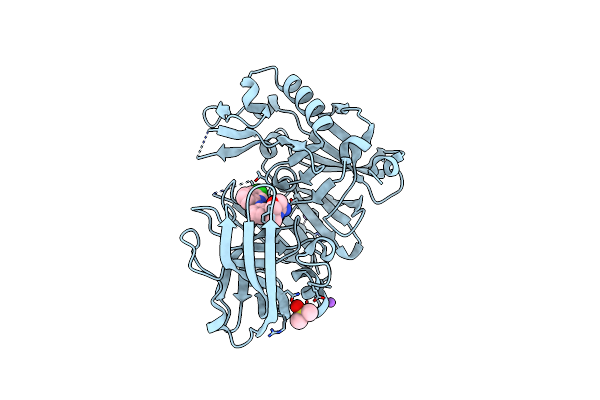
Crystal Structure Of Bace-1 In Complex With (4S)-4-[3-(5-Chloro-3-Pyridyl)Phenyl]-4-[4-(Difluoromethoxy)-3-Methyl-Phenyl]-5H-Oxazol-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-24 Classification: Hydrolase/Inhibitor Ligands: 5T6, DMS, NA |
 |
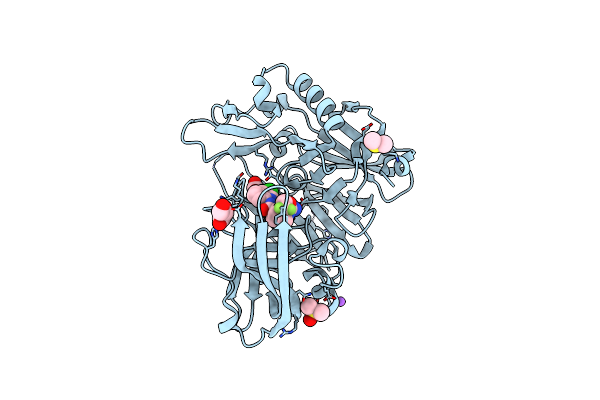
Crystal Structure Of Bace-1 In Complex With 5-[3-[(3-Chloro-8-Quinolyl)Amino]Phenyl]-5-Methyl-2,6-Dihydro-1,4-Oxazin-3-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-02-24 Classification: HYDROLASE |
 |
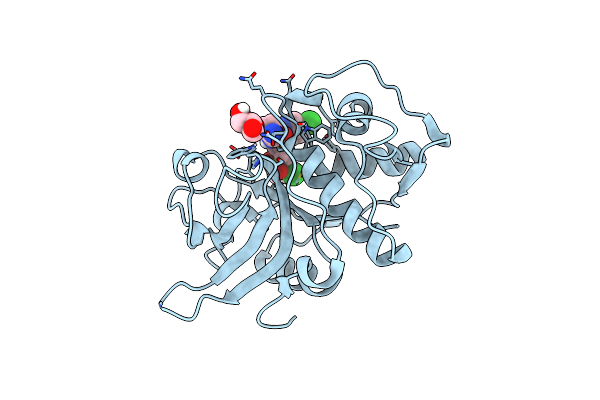
Crystal Structure Of Bace-1 In Complex With (1Sr,2Sr)-2-((R)-2-Amino-5,5-Difluoro-4-Methyl-5,6-Dihydro-4H-1,3-Oxazin-4-Yl)-N-(3-Chloroquinolin-8-Yl)Cyclopropanecarboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2016-02-24 Classification: HYDROLASE |
 |
Cathepsin L In Complex With (2S,4R)-4-(2-Chloro-4-Methoxy-Benzenesulfonyl)-1-[3-(5-Chloro-Pyridin-2-Yl)-Azetidine-3-Carbonyl]-Pyrrolidine-2-Carboxylic Acid (1-Cyano-Cyclopropyl)-Amide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2016-02-24 Classification: HYDROLASE Ligands: GOL, 5T9 |
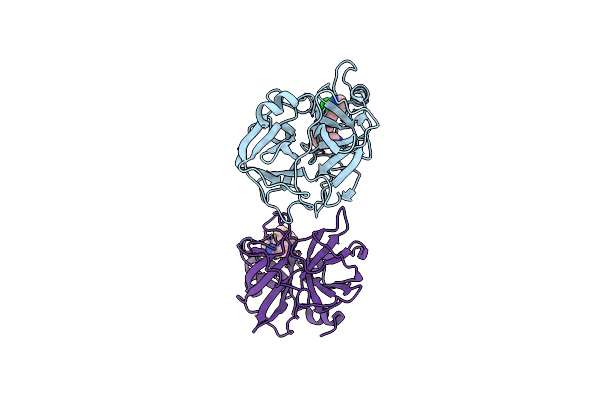 |
Tryptase B2 In Complex With 5-(3-Aminomethyl-Phenoxymethyl)-3-[3-(2-Chloro-Pyridin-3-Ylethynyl)-Phenyl]-Oxazolidin-2-One; Compound With Trifluoro-Acetic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-02-24 Classification: HYDROLASE Ligands: 5TA |
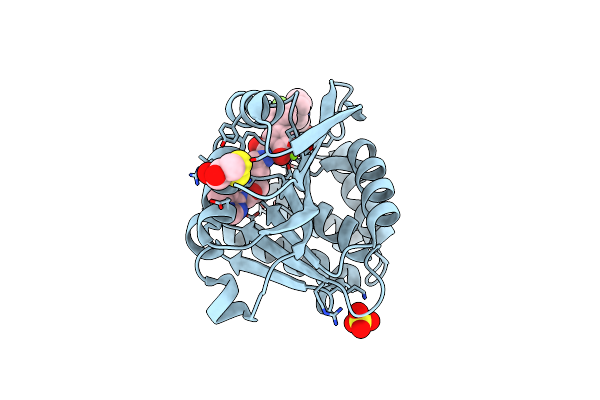 |
Rat Catechol O-Methyltransferase In Complex With The Bisubstrate Inhibitor 4'-Fluoro-4,5-Dihydroxy-Biphenyl-3-Carboxylic Acid {(E)-3-[(2S,4R,5R)-4-Hydroxy-5-(6-Methyl-Purin-9-Yl)-Tetrahydro-Furan-2-Yl]-Allyl}-Amide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, CL, SO4, LU1, DTD, NHE |
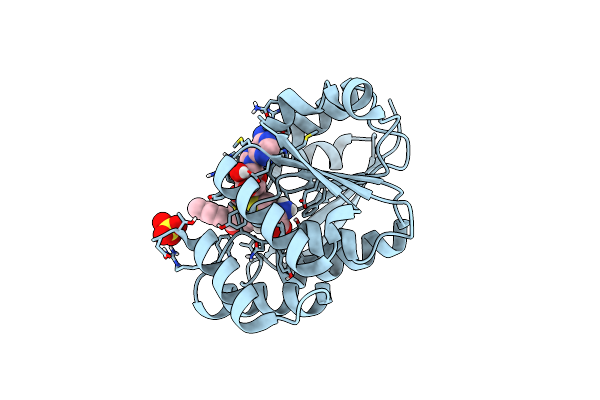 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, SO4, CL, TCW, SAM |
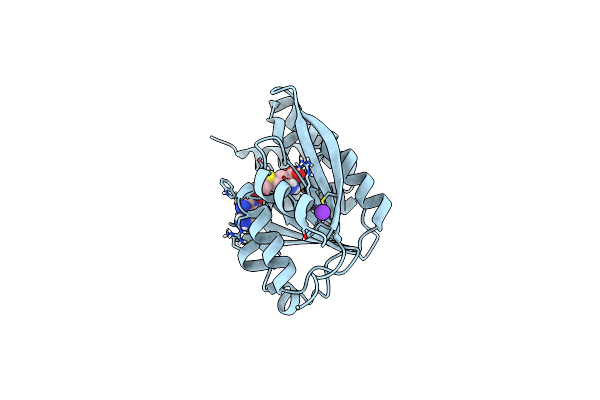 |
Crystal Structure Of A Sah-Bound Semi-Holo Form Of Rat Catechol-O-Methyltransferase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SAH, K |
 |
Rat Comt In Complex With A Fluorinated Desoxyribose-Containing Bisubstrate Inhibitor Avoids Hydroxyl Group
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-08-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, CL, 659, NHE, SO4 |
 |
Rat Comt In Complex With A Methylated Desoxyribose Bisubstrate-Containing Inhibitor Avoids Hydroxyl Group
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-08-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, CL, SO4, 662, NHE |

