Search Count: 213
 |
Structure Of Saro_1862, A Upf0261 Domain Protein From Novosphingobium Aromaticivorans With Bound Acetovanillone
Organism: Novosphingobium aromaticivorans dsm 12444
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-09-24 Classification: HYDROLASE Ligands: I75, CIT, NA |
 |
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.00 Å Release Date: 2025-09-03 Classification: IMMUNE SYSTEM Ligands: CL |
 |
In Situ Microed Structure Of Il-5 Activated Human Eosinophil Major Basic Protein-1
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.18 Å Release Date: 2025-09-03 Classification: IMMUNE SYSTEM Ligands: CL |
 |
In Situ Microed Structure Of Il-33 Activated Human Eosinophil Major Basic Protein-1
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.18 Å Release Date: 2025-09-03 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Organism: Tequatrovirus t4
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2025-05-14 Classification: VIRAL PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-10-16 Classification: RNA Ligands: K, NA |
 |
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.20 Å Release Date: 2024-09-25 Classification: IMMUNE SYSTEM Ligands: HOH |
 |
Cystathionine Gamma Lyase From Thermobifida Fusca In Internal Aldimine Form
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-07-03 Classification: LYASE Ligands: PEG, 1PE, MG |
 |
Parathyroid Hormone 1 Receptor Extracellular Domain Complexed With A Peptide Ligand Containing Beta-3-Homotryptophan
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-14 Classification: SIGNALING PROTEIN Ligands: EDO, ZN |
 |
Parathyroid Hormone 1 Receptor Extracellular Domain Complexed With A Peptide Ligand Containing (2-Naphthyl)-Beta-3-Homoalanine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-06-14 Classification: SIGNALING PROTEIN Ligands: EDO, ZN |
 |
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-05 Classification: HYDROLASE Ligands: CA |
 |
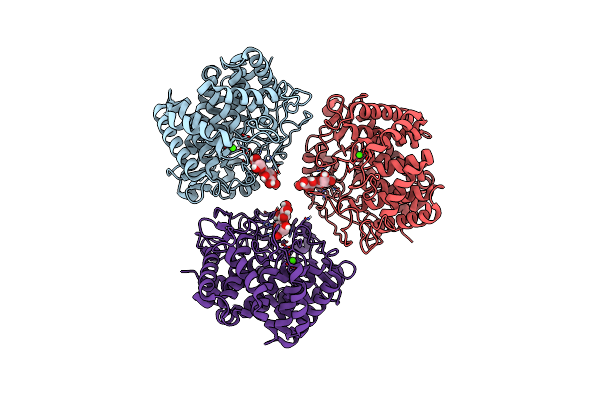
Crystal Structure Of A Celr Catalytic Domain Active Site Mutant With Bound Cellohexaose Substrate
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-05 Classification: HYDROLASE/SUBSTRATE Ligands: CA |
 |
Crystal Structure Of A Celr Catalytic Domain Active Site Mutant With Bound Cellobiose Product
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-05 Classification: HYDROLASE/PRODUCT Ligands: CA |
 |
Crystal Structure Of Saccharomyces Cerevisiae Altered Inheritance Rate Of Mitochondria Protein 18 (Aim18P) R123A Mutant
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-03-22 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of Saccharomyces Cerevisiae Altered Inheritance Rate Of Mitochondria Protein 46 (Aim46P)
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-22 Classification: UNKNOWN FUNCTION Ligands: AKG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: MVS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-01-18 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MVS, NA |
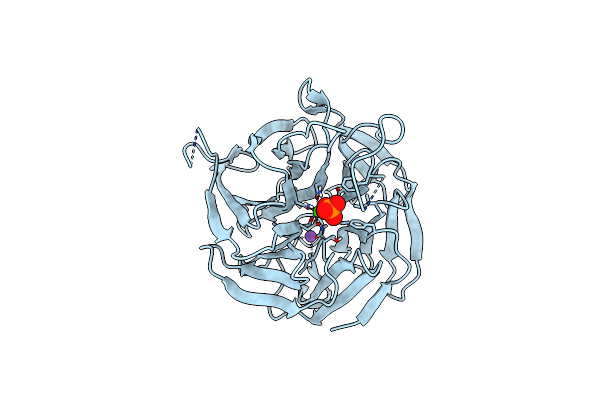 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound Phosphate
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: PO4, CA, NA |
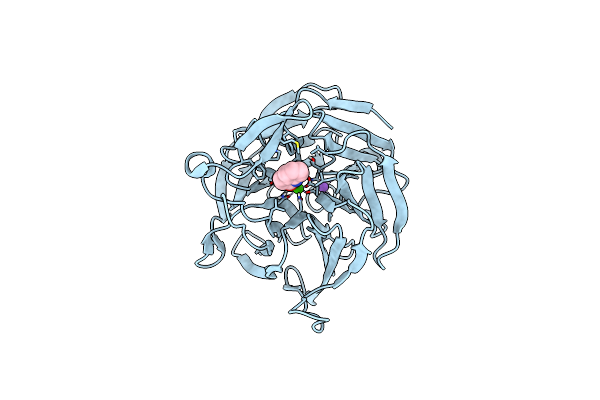 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound 2-Hydroxyquinoline
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: OCH, CA, NA |
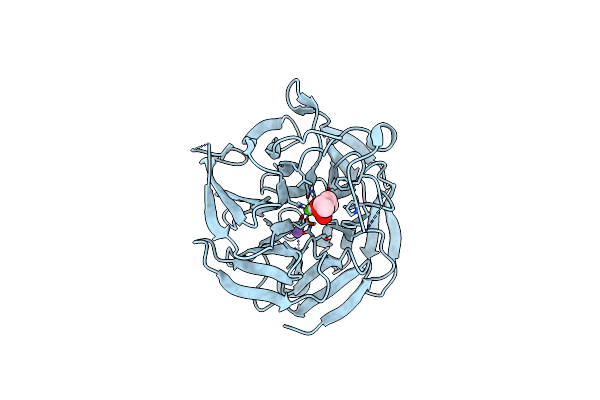 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound Tetrahedral Intermediate
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: CA, NA, SGU |

