Search Count: 16
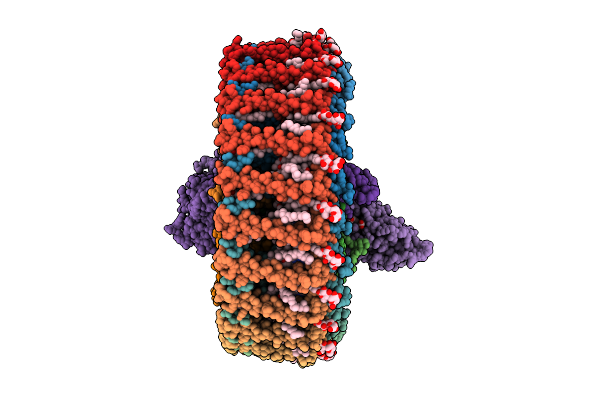 |
Cryo Em Structure Of Rc-Dlh Complex Model Ii From Gemmatimonas Groenlandica
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, LMT, MQ8, FE, CD4, CRT, PEX, HEC, V7N |
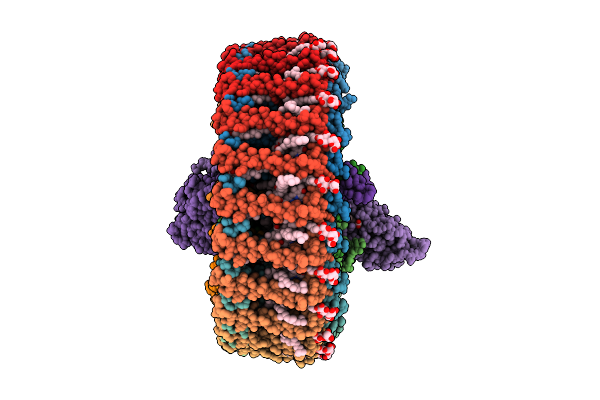 |
Cryo-Em Structure Of Rc-Dlh Complex Model I From Gem. Groenlandica Strain Tet16
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PHOTOSYNTHESIS Ligands: V7N, BCL, LMT, PEX, MQ8, CD4, BPH, FE, CRT, HEC |
 |
Bottom Cylinder Of High-Resolution Phycobilisome Quenched By Ocp (Local Refinement)
Organism: Synechocystis sp. pcc 6803
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PHOTOSYNTHESIS Ligands: CYC, 45D |
 |
Central Rod Disk In C1 Symmetry Of High-Resolution Phycobilisome Quenched By Ocp (Local Refinement)
Organism: Synechocystis sp. pcc 6803
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Top Cylinder Bound To Ocp From High-Resolution Phycobilisome Quenched By Ocp (Local Refinement)
Organism: Synechocystis sp. pcc 6803
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PHOTOSYNTHESIS Ligands: 45D, CYC |
 |
Organism: Synechocystis sp. pcc 6803
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PHOTOSYNTHESIS Ligands: CYC |
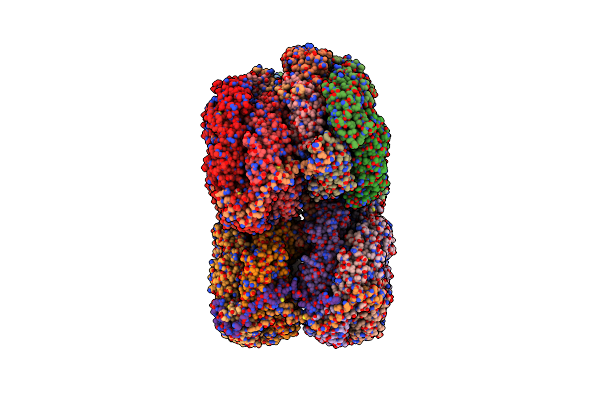 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC |
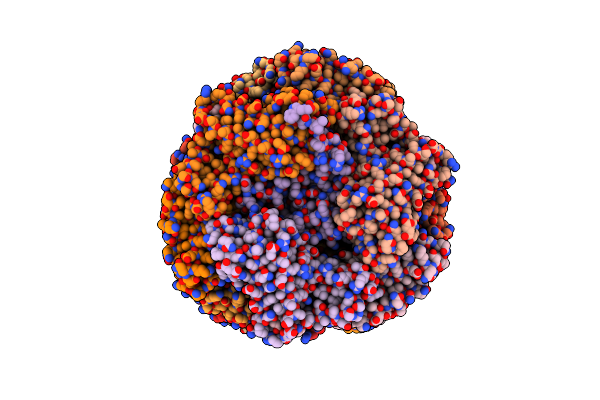 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC, 45D |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
B-Cylinder Of Synechocystis Pcc 6803 Phycobilisome, Complex With Ocp - Local Refinement
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC, 45D |
 |
T-Cylinder Of Synechocystis Pcc 6803 Phycobilisome, Complex With Ocp - Local Refinement
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: PHOTOSYNTHESIS Ligands: CYC, 45D |
 |
Cryo-Em Structure (Model_1A) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.4 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, 0V9, HEC, NDG, V75, CD4, PGW, MQ8, BPH, FE, CRT, V7B, UYH |
 |
Cryo-Em Structure (Model_2A) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.5 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, HEC, V75, NDG, 0V9, CD4, PGW, MQ8, V7B, BPH, FE, CRT, UYH |
 |
Cryo-Em Structure Of The Rc-Dlh Complex (Model_1B) From Gemmatimonas Phototrophica At 2.47 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, 0V9, HEC, V75, NDG, PGW, CD4, BPH, MQ8, FE, CRT, V7B, UYH |
 |
Cryo-Em Structure (Model_2B) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.44 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, HEC, NDG, V75, PGW, 0V9, CD4, BPH, MQ8, FE, CRT, V7B, UYH |

