Search Count: 17
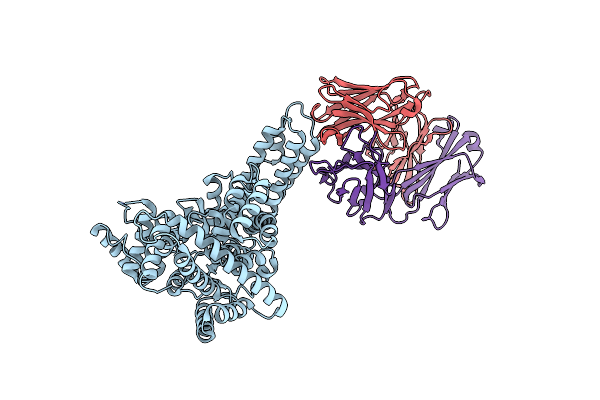 |
Novel Structural Insights For A Pair Of Monoclonal Antibodies Recognizing Non-Overlapping Epitopes Of The Glucosyltransferase Domain Of Clostridium Difficile Toxin B
Organism: Homo sapiens, Clostridioides difficile r20291
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-11 Classification: TOXIN/IMMUNE SYSTEM |
 |
Novel Structural Insights For A Pair Of Monoclonal Antibodies Recognizing Non-Overlapping Epitopes Of The Glucosyltransferase Domain Of Clostridium Difficile Toxin B
Organism: Clostridioides difficile, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2022-05-11 Classification: TOXIN/IMMUNE SYSTEM |
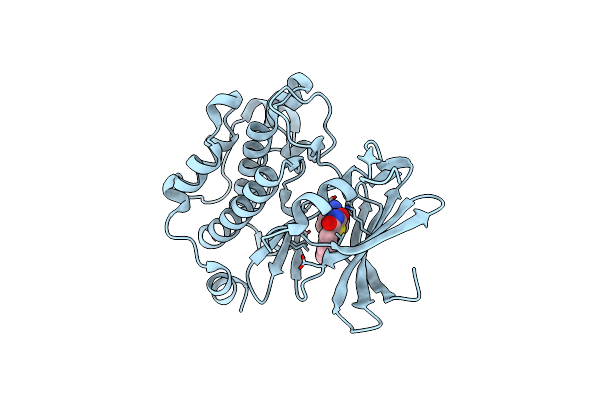 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0F5 |
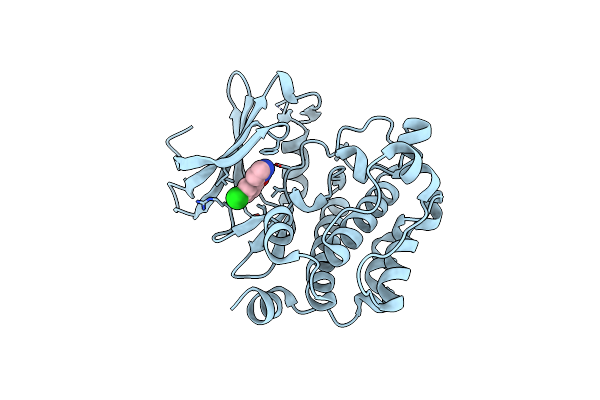 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0F9 |
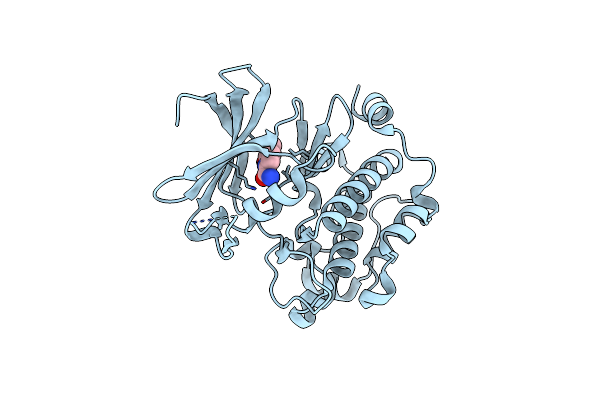 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0FK |
 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0FN |
 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0FO |
 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0FR, IMD |
 |
Exploitation Of Hydrogen Bonding Constraints And Flat Hydrophobic Energy Landscapes In Pim-1 Kinase Needle Screening And Inhibitor Design
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2012-03-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: IMD, 0FS |
 |
The Discovery Of Novel Benzofuran-2-Carboxylic Acids As Potent Pim-1 Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: IMD, UNJ |
 |
The Discovery Of Novel Benzofuran-2-Carboxylic Acids As Potent Pim-1 Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-05-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: IMD, UNE |
 |
The Discovery Of Novel Benzofuran-2-Carboxylic Acids As Potent Pim-1 Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-05-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: UNM, IMD |
 |
The Discovery Of Novel Benzofuran-2-Carboxylic Acids As Potent Pim-1 Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-05-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: UNQ, IMD |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1997-05-15 Classification: COMPLEX (CHEMOTAXIS/PEPTIDE) Ligands: ASP |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1997-04-21 Classification: CHEMOTAXIS |
 |
The Three-Dimensional Structure Of The Ligand-Binding Domain Of A Wild-Type Bacterial Chemotaxis Receptor
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 1994-12-20 Classification: CHEMOTAXIS |
 |
The Three-Dimensional Structure Of The Ligand-Binding Domain Of A Wild-Type Bacterial Chemotaxis Receptor
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-12-20 Classification: CHEMOTAXIS Ligands: ASP |

