Search Count: 91
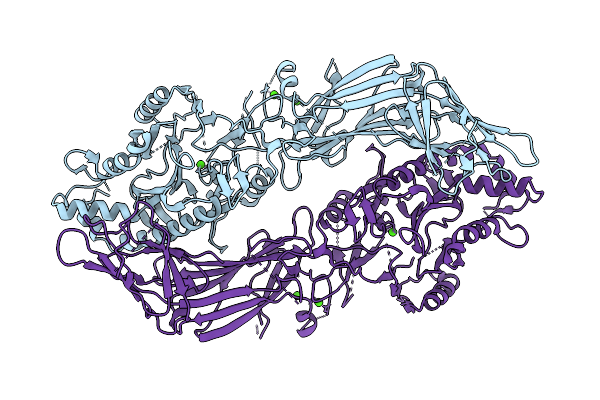 |
Cryoem Structure Of Human Peptidylarginine Deiminase Type 4 (Pad4) In 10 Mm Calcium
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: CA |
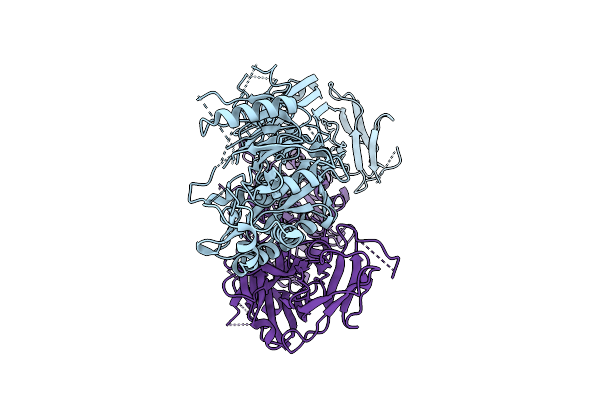 |
Cryoem Structure Of Human Peptidylarginine Deiminase Type 4 (Pad4) In Complex With Heparin Oligomer (20 Subunits).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
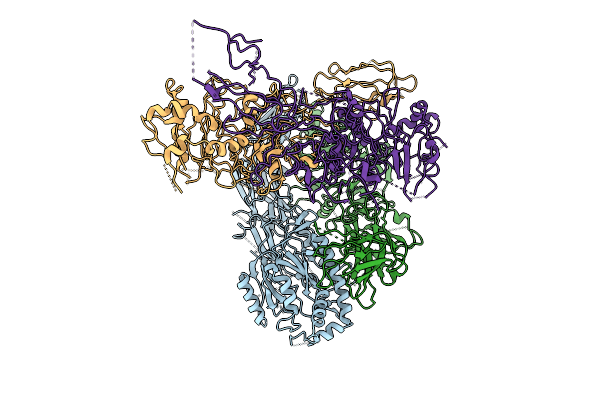 |
Cryoem Structure Of Human Peptidylarginine Deiminase Type 4 (Pad4) In Complex With Heparin Oligomer (12 Subunits)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
 |
Organism: Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRUS LIKE PARTICLE Ligands: CO |
 |
Structure Of The Co(Ii) Triggered Trap (S33Hk35H) Protein Cage (Dextro Form)
Organism: Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRUS LIKE PARTICLE Ligands: CO |
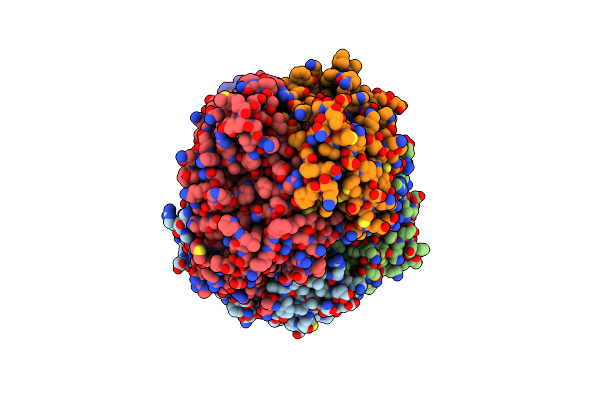 |
Cryo-Em Structure Of Dhs-Erk2 Complex With 1:1 Stoichiometry Refined In C1 Symmetry
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: TRANSLATION |
 |
Conserved Structures And Dynamics In 5-Proximal Regions Of Betacoronavirus Rna Genomes
Organism: Human coronavirus oc43
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Conserved Structures And Dynamics In 5-Proximal Regions Of Betacoronavirus Rna Genomes
Organism: Rousettus bat coronavirus gccdc1
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Conserved Structures And Dynamics In 5-Proximal Regions Of Betacoronavirus Rna Genomes
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Conserved Structures And Dynamics In 5-Proximal Regions Of Betacoronavirus Rna Genomes
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
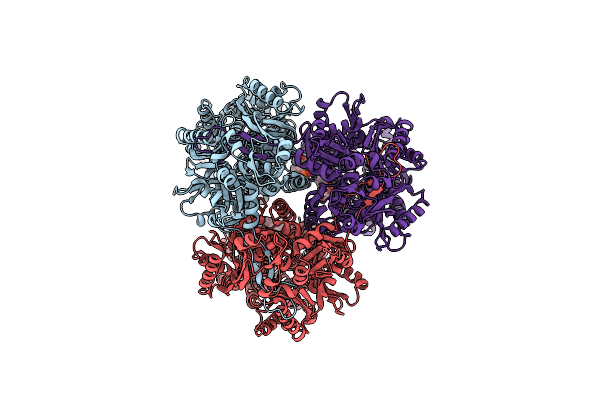 |
Single Particle Cryo-Em Co-Structure Of Klebsiella Pneumoniae Acrb With The Bdm91288 Efflux Pump Inhibitor At 2.97 Angstrom Resolution
Organism: Klebsiella pneumoniae subsp. pneumoniae dsm 30104 = jcm 1662 = nbrc 14940
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: WQW, WR6, PG8 |
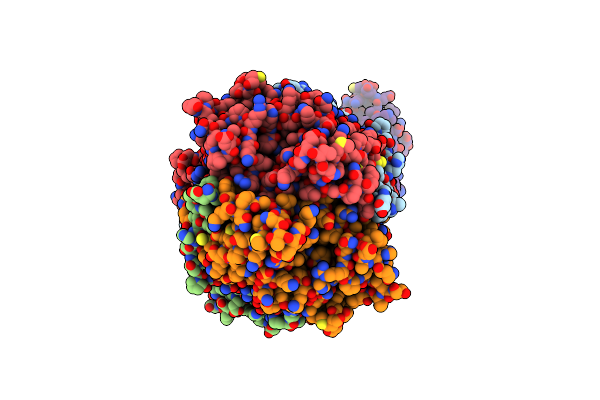 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSFERASE Ligands: NAD, SPD |
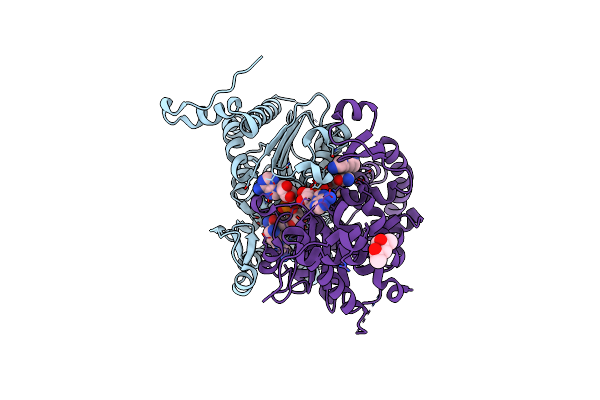 |
Crystal Structure Of Human Deoxyhypusine Synthase Variant K329A In Complex With Nad And Spd
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-04-05 Classification: TRANSFERASE Ligands: SPD, EDO, NAD, MRD |
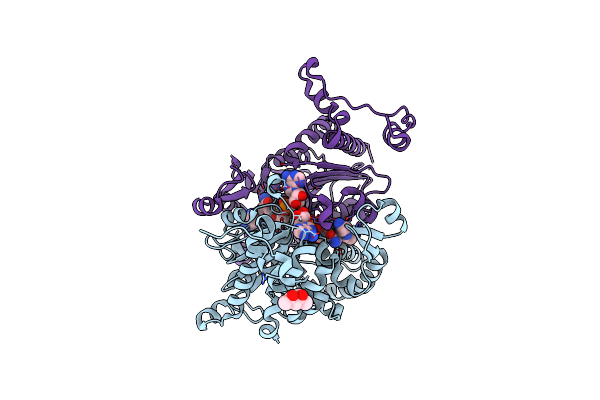 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-04-05 Classification: TRANSFERASE Ligands: 13D, MRD, NAD, SPD |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: TRANSLATION Ligands: SF4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: TRANSLATION Ligands: SF4, 5AD |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: TRANSLATION |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: TRANSLATION Ligands: SF4, SAM |
 |
Crystal Structure Of Endoplasmic Reticulum Aminopeptidase 2 (Erap2) Complex With A Highly Selective And Potent Small Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-07-20 Classification: IMMUNE SYSTEM,HYDROLASE/INHIBITOR Ligands: ZN, GIY, EPE, MES, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: EDO, ACT, OXM |

