Search Count: 121
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Sp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.65 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-06-05 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Sp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.41 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2024-06-05 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, CA, NA, PG4 |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: ASP, PG4, PGE, P6G, SAH, CA, NA |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, NA, CA, PG4, P6G |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, CA, PGE |
 |
Aeronamide N-Methyltransferase, Aere, Bound To Modified Peptide Substrate, Aera-Dl,34
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-03-30 Classification: TRANSFERASE/BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Bovine Lactoperoxidase With Hydrogen Peroxide Trapped Between Heme Iron And His109 At 1.69 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, GOL, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of Ternary Complexes Of Lactoperoxidase With Hydrogen Peroxide At 1.70 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Hydrogen Peroxide At 1.77A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-12-16 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, GOL, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of The Complex Of Goat Lactoperoxidase With Hypothiocyanite And Hydrogen Peroxide At 1.79 A Resolution.
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-01-22 Classification: OXIDOREDUCTASE Ligands: NAG, HEM, CA, IOD, EDO, PEO, OSM, SCN, GOL |
 |
Crystal Structure Of Hydrogen Peroxide Bound Bovine Lactoperoxidase At 2.3 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-08-28 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, PEO |
 |
Crystal Structure Of Peptidyl-Trna Hydrolase With Multiple Sodium And Chloride Ions At 1.08 A Resolution.
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: CL, MOH, NA |
 |
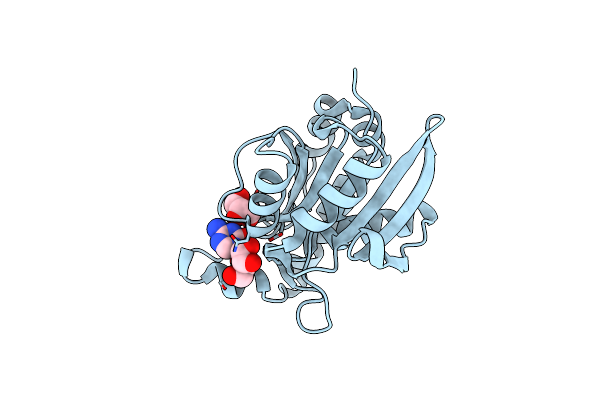
Crystal Structure Of Peptidyl-Trna Hydrolase From A Gram-Positive Bacterium, Streptococcus Pyogenes At 2.19 Angstrom Resolution Shows The Closed Structure Of The Substrate Binding Cleft
Organism: Streptococcus pyogenes nz131
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-08-06 Classification: HYDROLASE |
 |
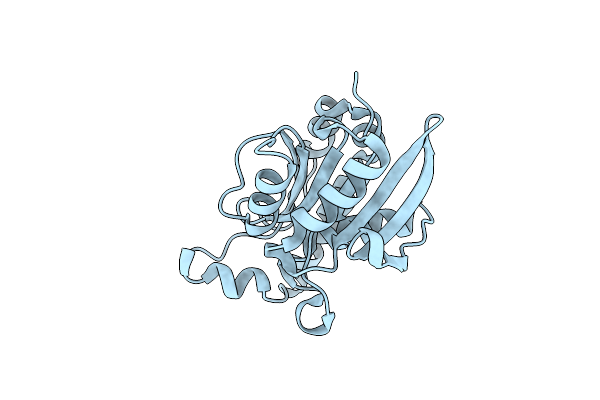
Crystal Structure Of Peptidyl-Trna Hydrolase From Pseudomonas Aeruginosa With 5-Azacytidine At 1.89 Angstrom Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: 5AE, GOL |
 |
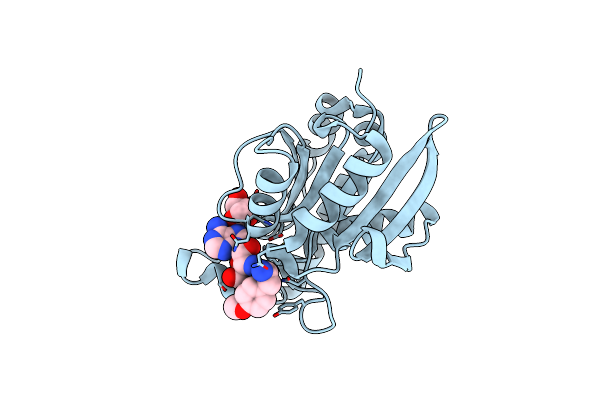
Crystal Structure Of Peptidyl-Trna Hydrolase From Pseudomonas Aeruginosa At 1.5 Angstrom Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-05-28 Classification: HYDROLASE |
 |
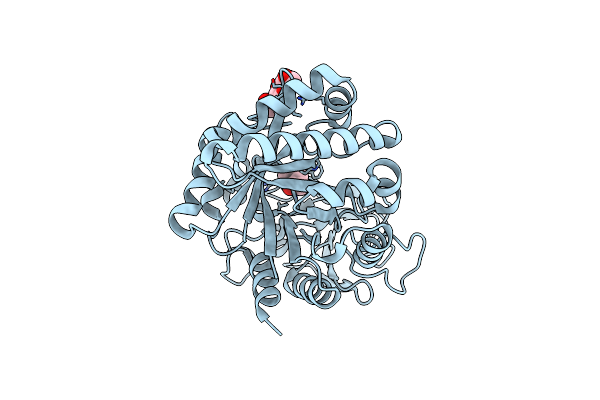
Crystal Structure Of The Complex Of Peptidyl-Trna Hydrolase From Pseudomonas Aeruginosa With Amino Acyl-Trna Analogue At 1.77 Angstrom Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: GOL, 3NZ |
 |
Crystal Structure Of The Complex Of Buffalo Signalling Protein Spb-40 With 4-N-Trimethylaminobutyraldehyde At 1.79 Angstrom Resolution
Organism: Bubalus bubalis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-05-21 Classification: SIGNALING PROTEIN Ligands: NAG, 2ZO |
 |
Crystal Structure Of Mammalian Peptidoglycan Recognition Protein Pgrp-S With Paranitrophenyl Palmitate And N-Acetyl Glucosamine At 2.09 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2014-05-21 Classification: IMMUNE SYSTEM Ligands: PNR, NAG, GOL, TLA |
 |
Crystal Structure Of Chitinase D From Serratia Proteamaculans In Complex With N-Acetyl Glucosamine At 1.93 Angstrom Resolution
Organism: Serratia proteamaculans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: NAG, ACT, GOL |

