Search Count: 19
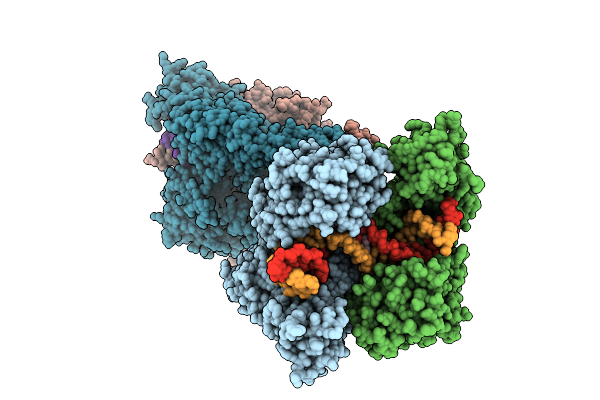 |
Crystal Structure Of Topoisomerase Iv From Klebsiella Pneumoniae In Complex With Dna And Bwc0977, A Dual-Targeting Broad-Spectrum Novel Bacterial Topoisomerase Inhibitor.
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: A1L5V |
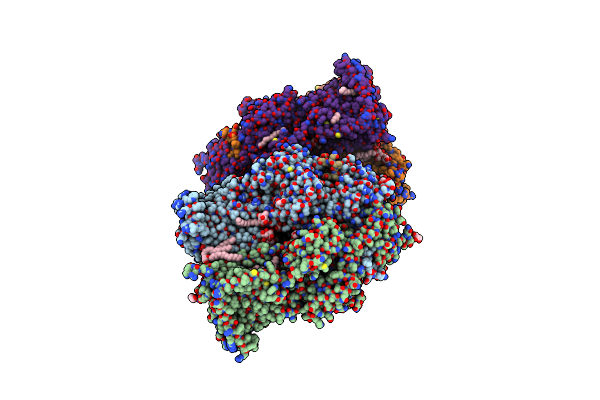 |
Crystal Structure Of Multidrug Efflux Transporter Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-22 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY, GOL |
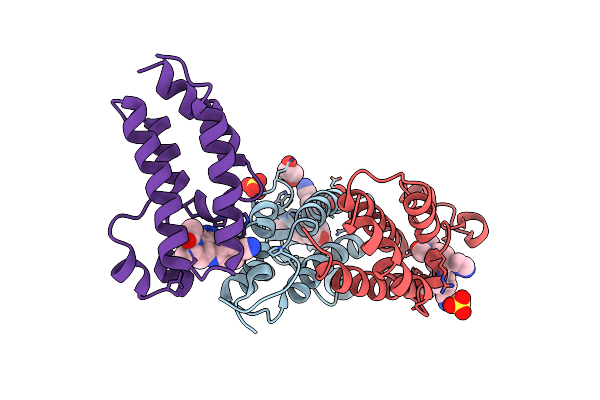 |
Crystal Structure Of The Human Brd2 Bd1 Bromodomain In Complex With A Tetrahydroquinoline Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-13 Classification: Transcription/Inhibitor Ligands: G7V, SO4, EDO, 03S |
 |
Crystal Structure Of The Human Brd2 Bd2 Bromodimain In Complex With A Tetrahydroquinoline Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-11-13 Classification: Transcription/Inhibitor Ligands: G7V, EDO |
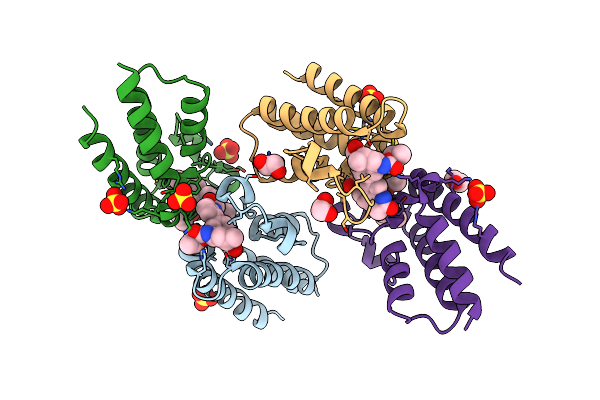 |
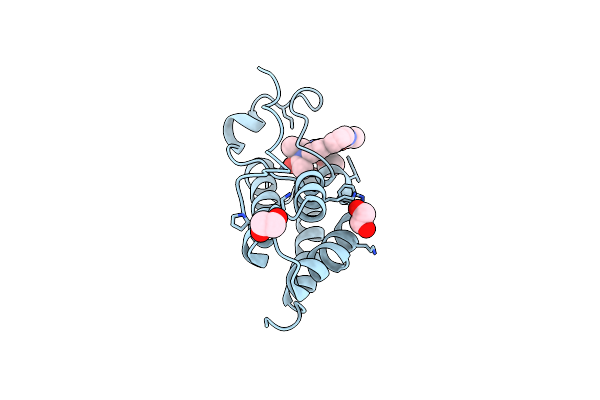
Crystal Structure Of The Second Bromodomain Of Human Brd2 In Complex With A Tetrahydroquinoline Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-02-28 Classification: TRANSCRIPTION Ligands: SO4, EDO, 8J2 |
 |
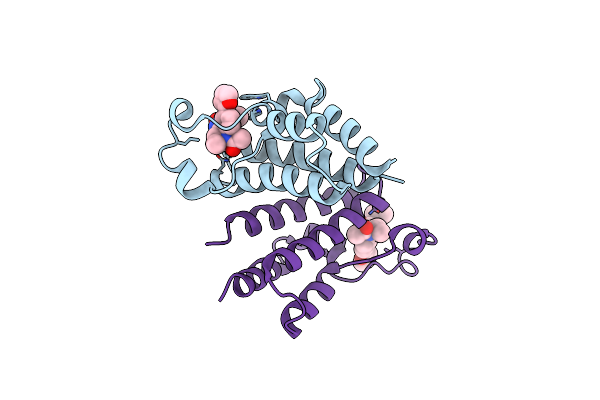
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With A Tetrahydroquinoline Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-02-28 Classification: TRANSCRIPTION Ligands: 8J2, EDO |
 |
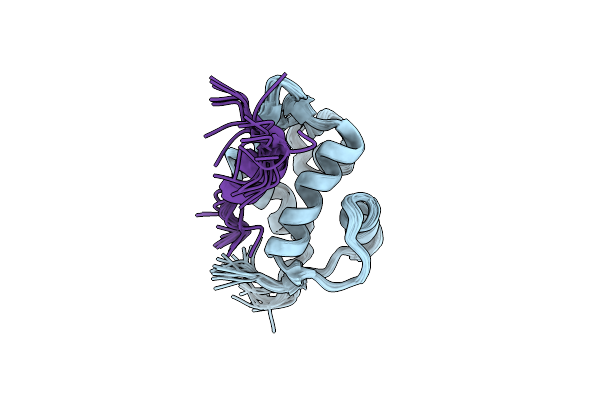
Crystal Structure Of The Second Bromodomain Of Human Brd2 In Complex With A Tetrahydroquinoline Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: 5P4 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: Cell Cycle/Antitumor Protein |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: SIGNALING PROTEIN Ligands: 44Z |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: SIGNALING PROTEIN Ligands: 48M |
 |
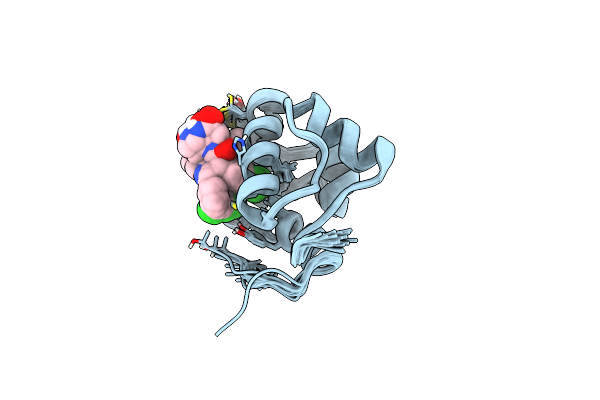
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: SIGNALING PROTEIN Ligands: 48L |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: SIGNALING PROTEIN Ligands: 49H |
 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H182A Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H432N Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H432N_Glu Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase 1 H432R Mutant (Scan1 Mutant)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
 |
Crystal Structure Of The Fk506 Binding Domain Of Plasmodium Vivax Fkbp35 In Complex With Fk506
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2010-06-16 Classification: ISOMERASE Ligands: FK5 |
 |
Plasmodium Falciparum Putative Fk506-Binding Protein Pfl2275C, C-Terminal Tpr-Containing Domain
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2006-01-24 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

