Search Count: 26
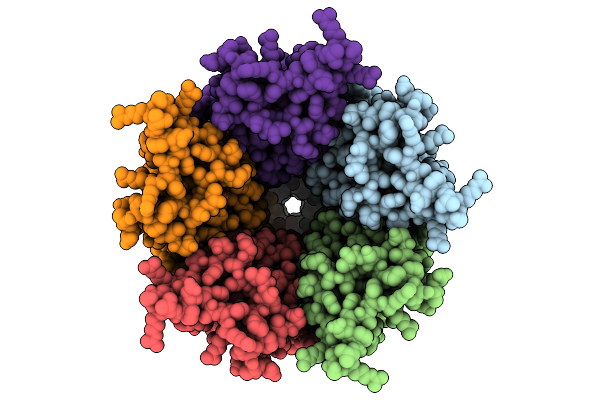 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiii Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
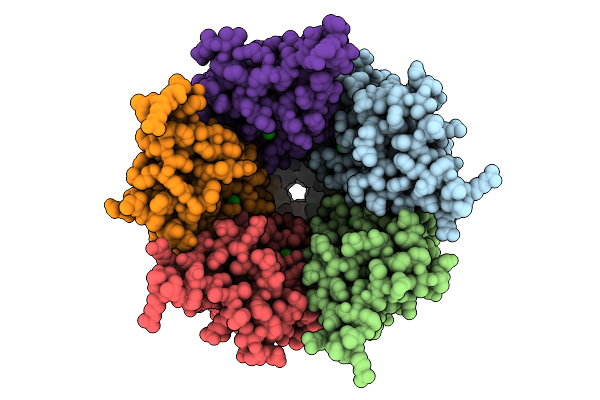 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
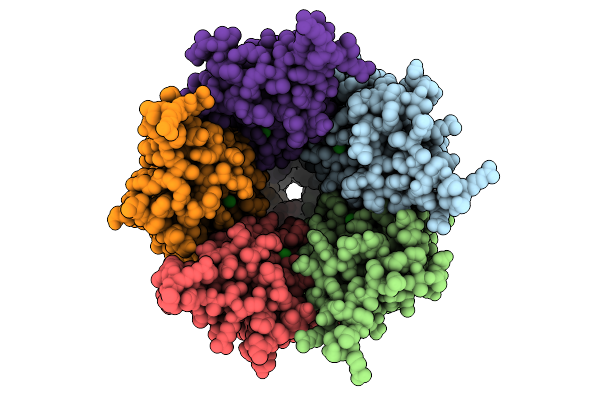 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
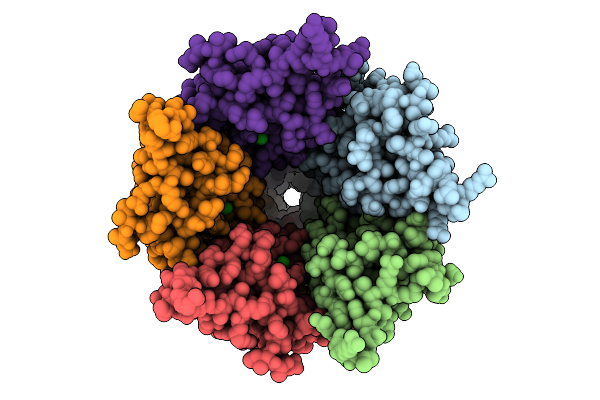 |
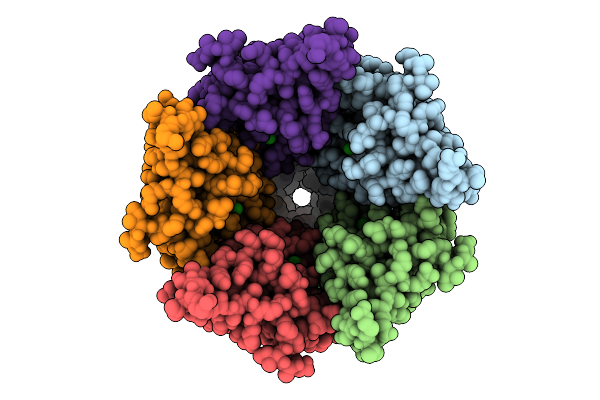
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
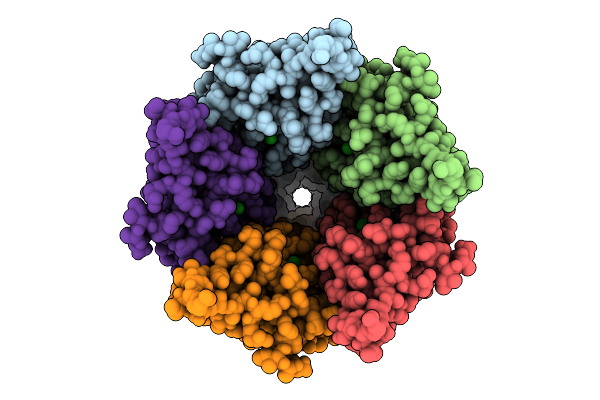
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
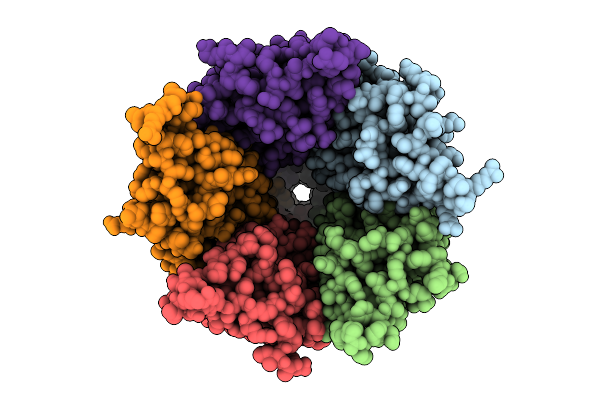
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ooooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
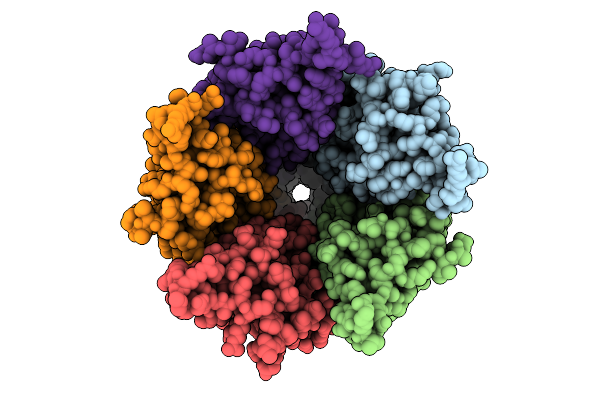
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iioio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic F116A Mutant At Ph 2.5
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic Y251A Mutant At Ph 2.5 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: CHAPERONE |
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Closed State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Intermediate State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Open State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: PLC |
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Crystal Structure Of An Adenylyl Cyclase Ma1120-Cat In Complex With Gtp And Calcium From Mycobacterium Avium
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Release Date: 2016-08-10 Classification: LYASE Ligands: CA, GTP, CL |
 |
Crystal Structure Of Triple Mutant (Kda To Egy) Of Adenylyl Cyclase Ma1120 From Mycobacterium Avium In Complex With Gtp And Calcium Ion
Organism: Mycobacterium avium subsp. avium 10-9275
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-08-10 Classification: LYASE Ligands: GTP, CA |

