Search Count: 20
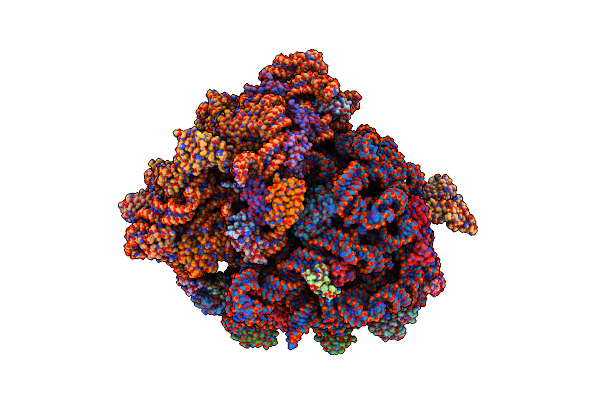 |
Molecular Model Of The Cryo-Em Structure Of 70S Ribosome In Complex With Peptide Deformylase And Trigger Factor
Organism: Escherichia coli (strain k12), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: RIBOSOME |
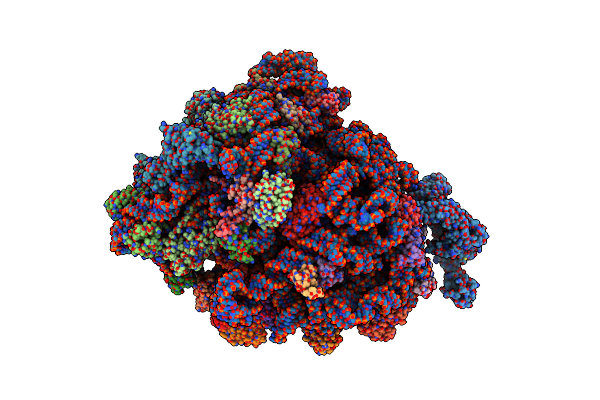 |
Molecular Model Of The Cryo-Em Structure Of 70S Ribosome In Complex With Peptide Deformylase, Trigger Factor, And Methionine Aminopeptidase
Organism: Escherichia coli (strain k12), Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:4.10 Å Release Date: 2021-04-07 Classification: RIBOSOME |
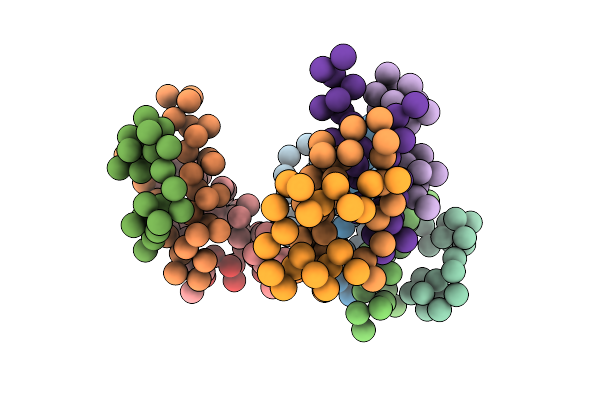 |
Crystal Structure Of Insulin Hexamer Fitted Into Cryo Em Density Map Where Each Dimer Was Kept As Rigid Body
|
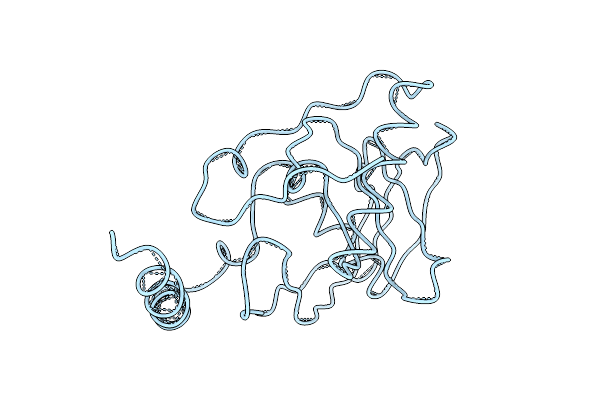 |
E. Coli Peptide Deformylase Crystal Structure Fitted Into The Cryo-Em Density Map Of E. Coli 70S Ribosome In Complex With Peptide Deformylase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
E. Coli Methionine Aminopeptidase Crystal Structure Fitted Into The Cryo-Em Density Map Of E. Coli 70S Ribosome In Complex With Methionine Aminopeptidase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Peptide Deformylase And Methionine Aminopeptidase Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Methionine Aminopeptidase Enzyme And Chaperone Trigger Factor Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli k-12, Thermotoga maritima msb8
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Peptide Deformylase Enzyme And Chaperone Trigger Factor Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli h591
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
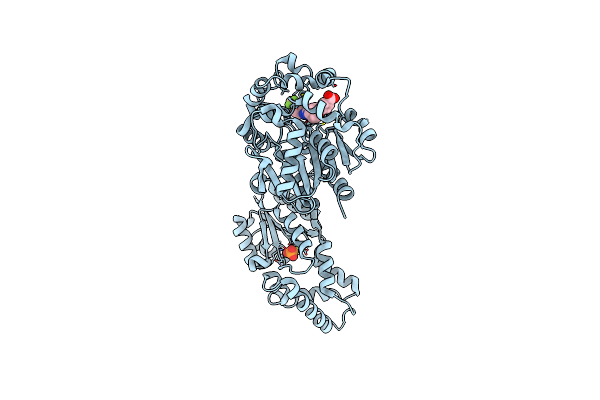 |
Crystal Structure Of Human Soluble Epoxide Hydrolase Complexed With Trans-4-[4-(3-Trifluoromethoxyphenyl-L-Ureido)-Cyclohexyloxy]-Benzoic Acid.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2018-02-07 Classification: hydrolase/hydrolase inhibitor Ligands: PO4, MG, BXV, CL |
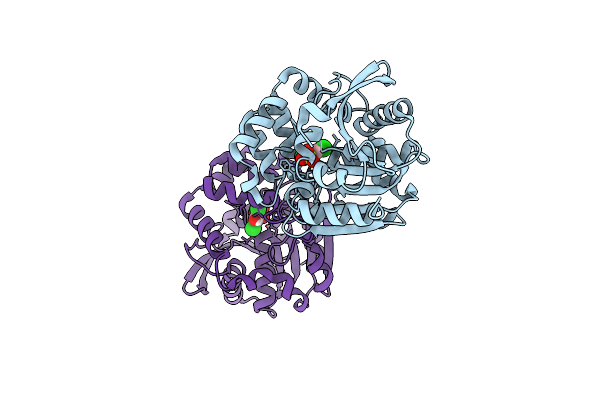 |
Crystal Structure Of Hsad Bound To 3,5-Dichloro-4-Hydroxybenzenesulphonic Acid
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2017-04-05 Classification: HYDROLASE Ligands: 6OR |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-04-05 Classification: HYDROLASE Ligands: 6OT, PO4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-04-05 Classification: HYDROLASE Ligands: FGZ |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2015-07-08 Classification: LYASE Ligands: NAG, DMS, 3P9 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-07-08 Classification: LYASE Ligands: EDO |
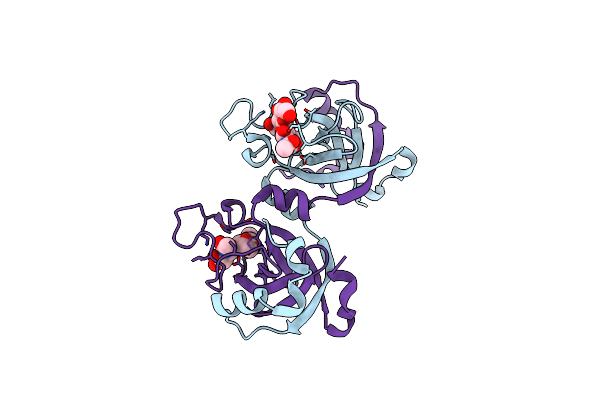 |
Stationary Phase Survival Protein Yuic From B.Subtilis Complexed With Reaction Product
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-07-08 Classification: LYASE Ligands: 3QL |
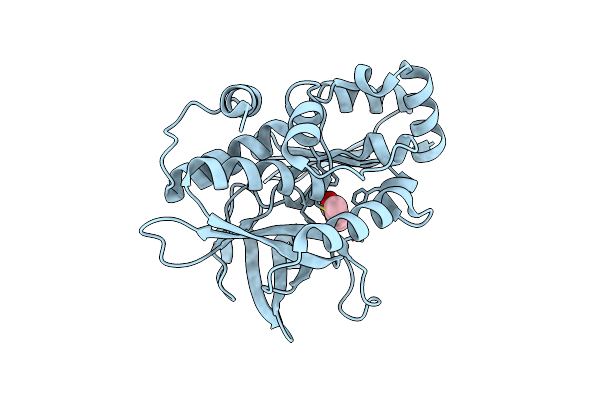 |
Crystal Structure Of The Covalent Adduct Formed Between Mycobacterium Marinum Aryalamine N-Acetyltransferase And Phenyl Vinyl Ketone A Derivative Of Piperidinols
Organism: Mycobacterium marinum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-01-16 Classification: TRANSFERASE Ligands: P18 |
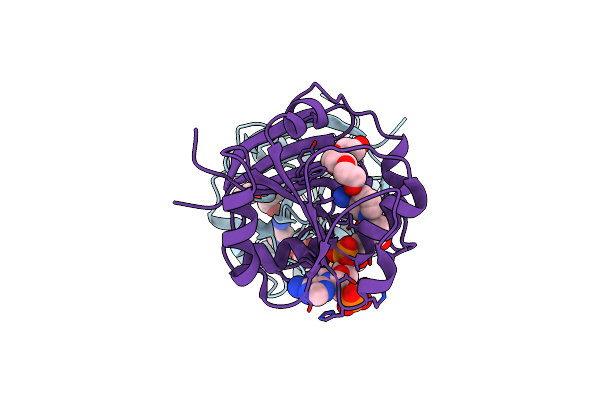 |
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: PG4, NDP, NA |
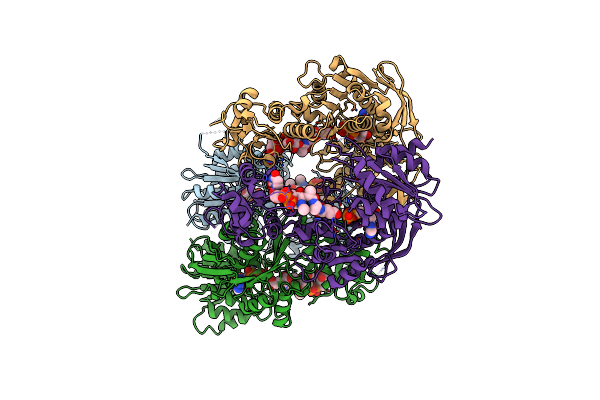 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-08-18 Classification: LIGASE Ligands: ADP, MG, UAG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-12-15 Classification: LIGASE Ligands: UAG, MG |
 |
X-Ray Crystallographic Structure Of Pseudomonas Aeruginosa Arylamine N-Acetyltransferase
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-08-03 Classification: TRANSFERASE Ligands: SO4 |

