Search Count: 52
 |
Crystal Structure Of Monomeric Plp-Dependent Transaminase From Desulfobacula Toluolica In F 41 3 2 Space Group
Organism: Desulfobacula toluolica
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-09-18 Classification: TRANSFERASE |
 |
Crystal Structure Of Monomeric Plp-Dependent Transaminase From Desulfobacula Toluolica In P 21 21 21 Space Group
Organism: Desulfobacula toluolica
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-09-18 Classification: TRANSFERASE Ligands: PEG, GOL |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Blastococcus Saxobsidens In Pmp Form
Organism: Blastococcus saxobsidens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: PMP, EDO, NA |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis In The Holo Form Obtained At Ph 7.0
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: PLP |
 |
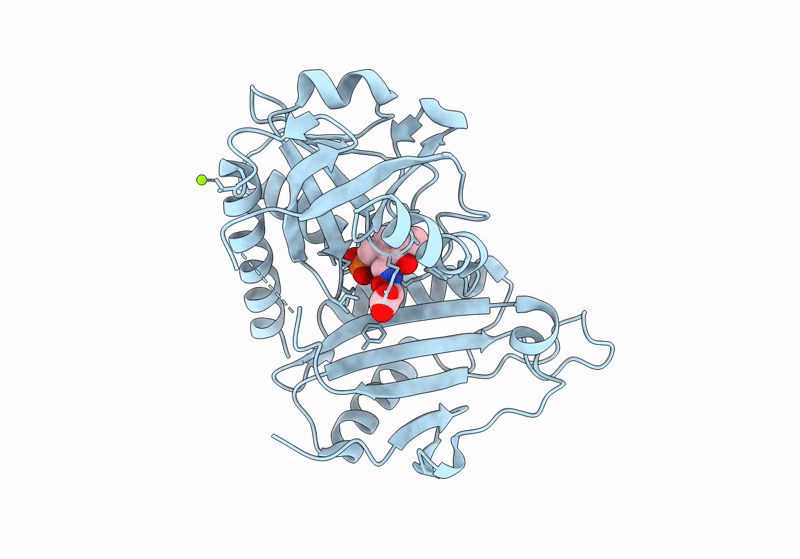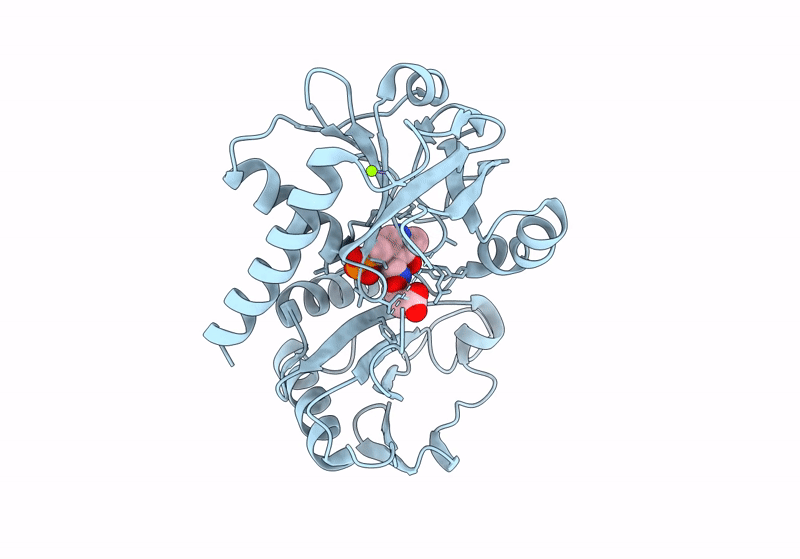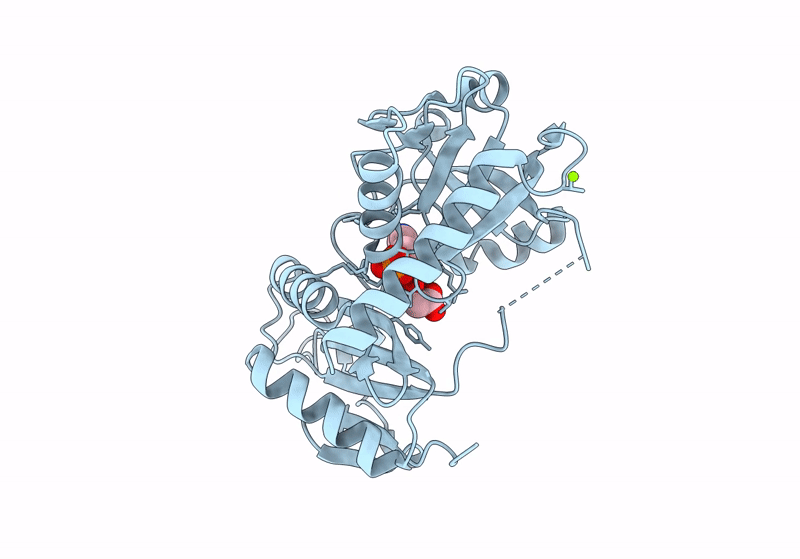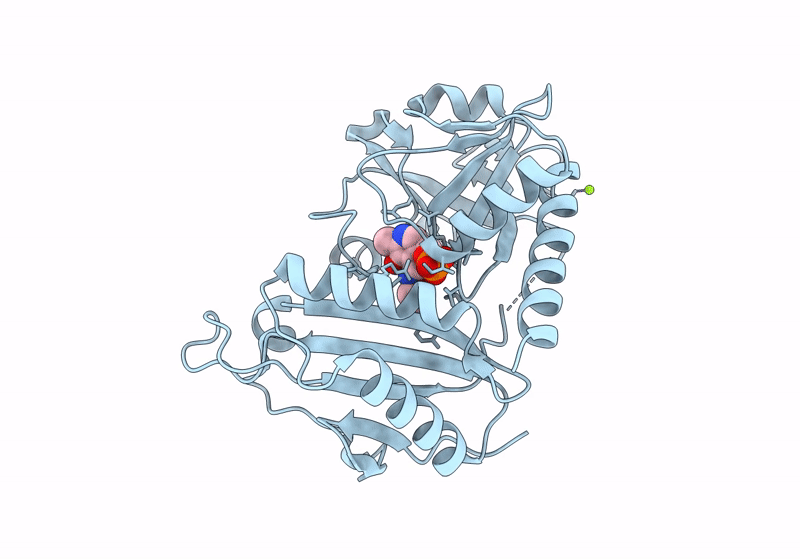
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis (Apo Form) After 15 Sec Of Soaking With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: ACT, ZXN |
 |
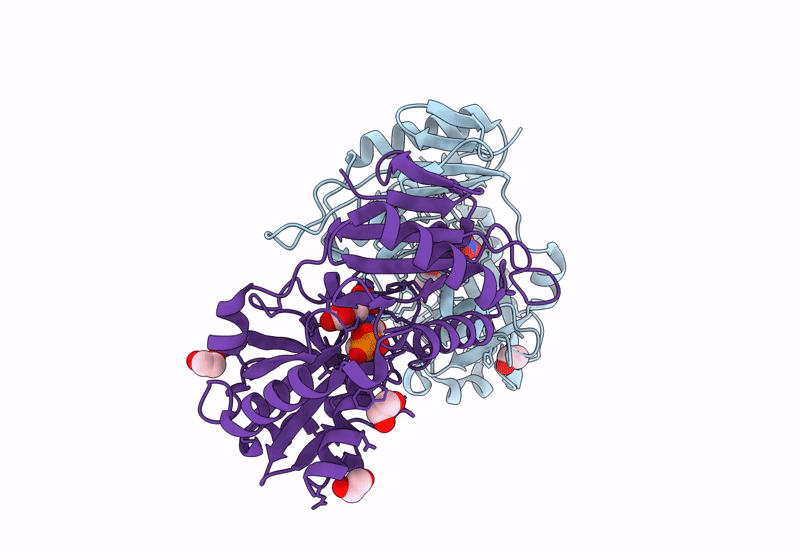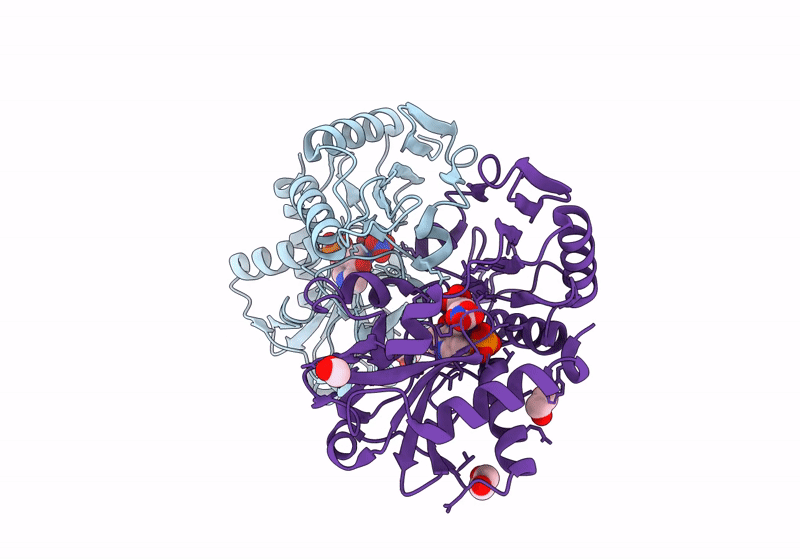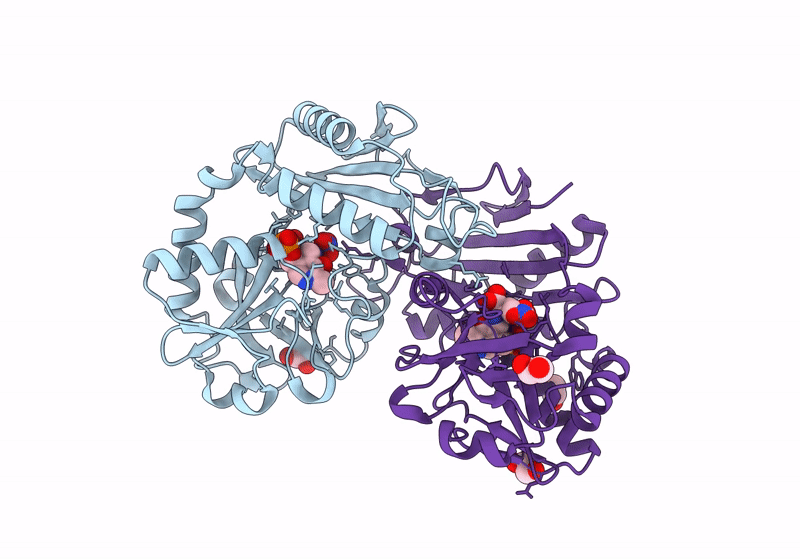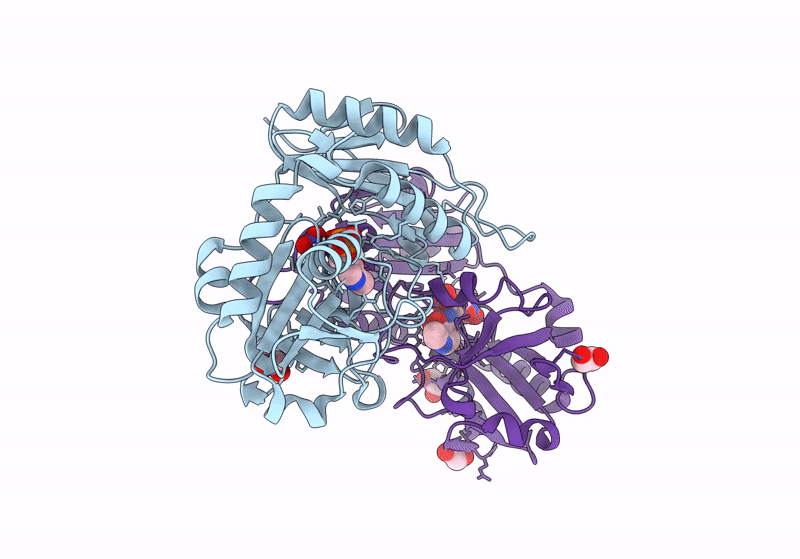
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Complexed With 3-Aminooxypropionic Acid
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: OCF, MG |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Point Mutant E113A Complexed With D-Glutamate
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: EDO, NO3, PMP, PW0 |
 |
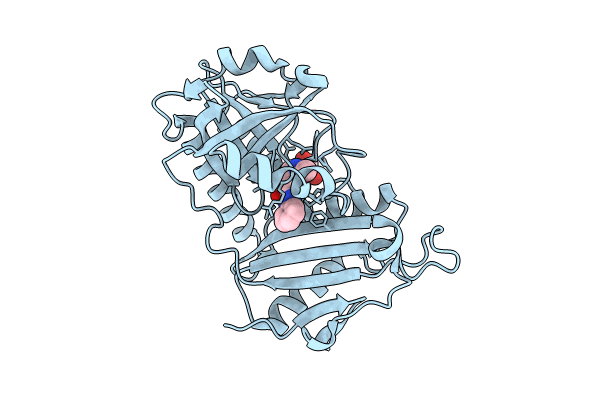
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I (Holo Form)
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I Complexed With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: ZXN, GOL |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Blastococcus Saxobsidens In Holo Form With Plp
Organism: Blastococcus saxobsidens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Blastococcus Saxobsidens Complexed With Phenylhydrazine And In Its Apo Form
Organism: Blastococcus saxobsidens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-25 Classification: TRANSFERASE Ligands: ZXN |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Point Mutant R88L
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: PLP, PEG |
 |
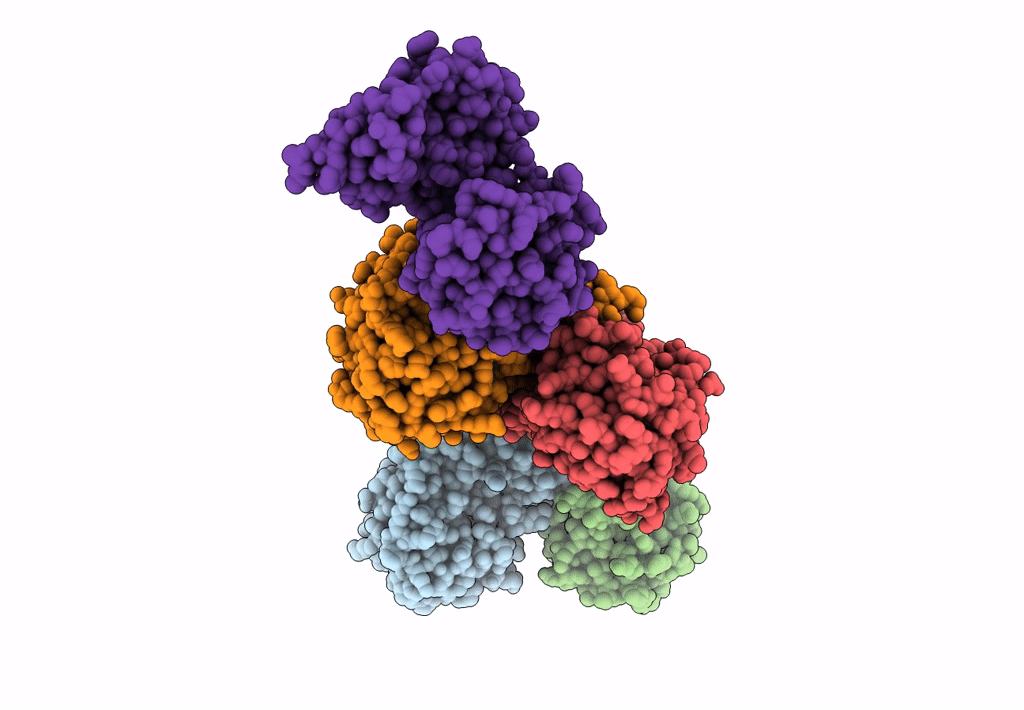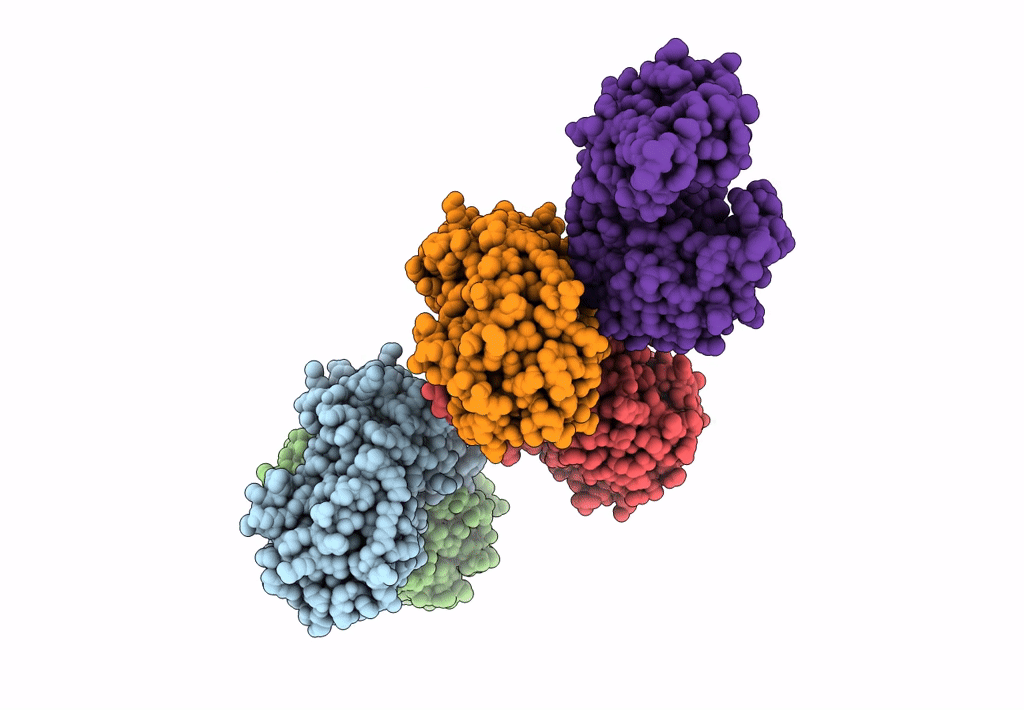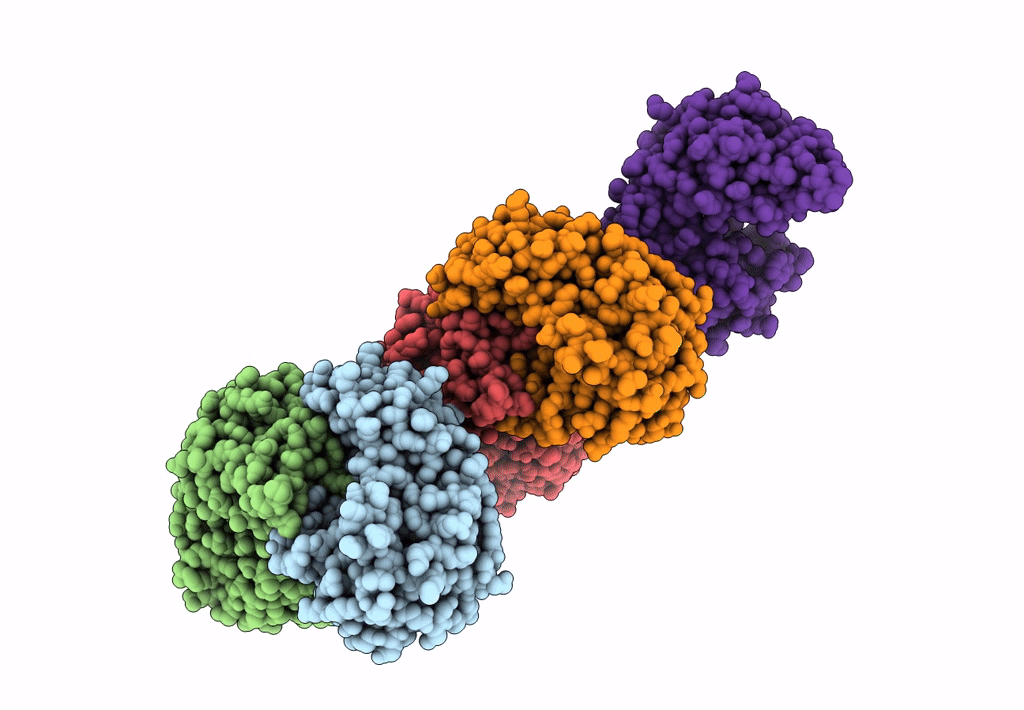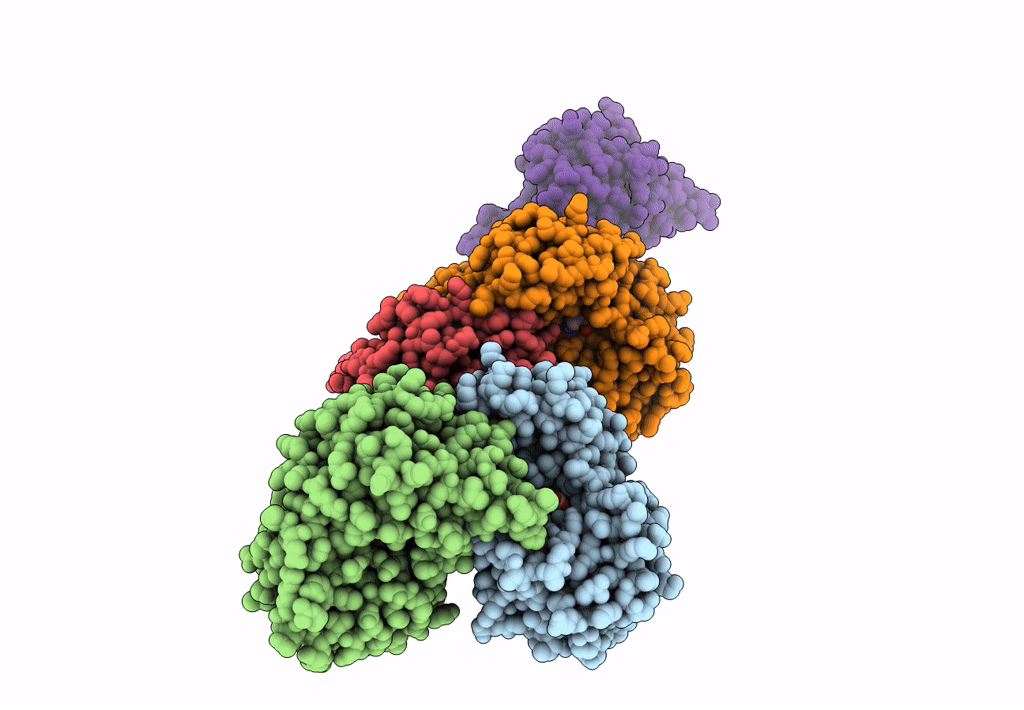
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Point Mutant E113A
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Point Mutant E113A Complexed With 3-Aminooxypropionic Acid
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: OCF |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Complexed With D-Cycloserine
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: LCS, PMP, M7L |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiens Complexed With 3-Aminooxypropionic Acid
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: OCF, EDO, PLP |
 |
Crystal Structure Of D-Amino Acid Aminotrensferase From Aminobacterium Colombiense Complexed With D-Glutamate
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: PW0, PMP |
 |
Crystal Structure Of D-Amino Acid Aminotrensferase From Haliscomenobacter Hydrossis Complexed With D-Cycloserine
Organism: Haliscomenobacter hydrossis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: GOL, LCS |
 |
Crystal Structure Of D-Aminoacid Transaminase From Haliscomenobacter Hydrossis In Its Intermediate Form
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: SO4, MG |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense In Holo Form With Plp
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: PLP |

