Search Count: 23
 |
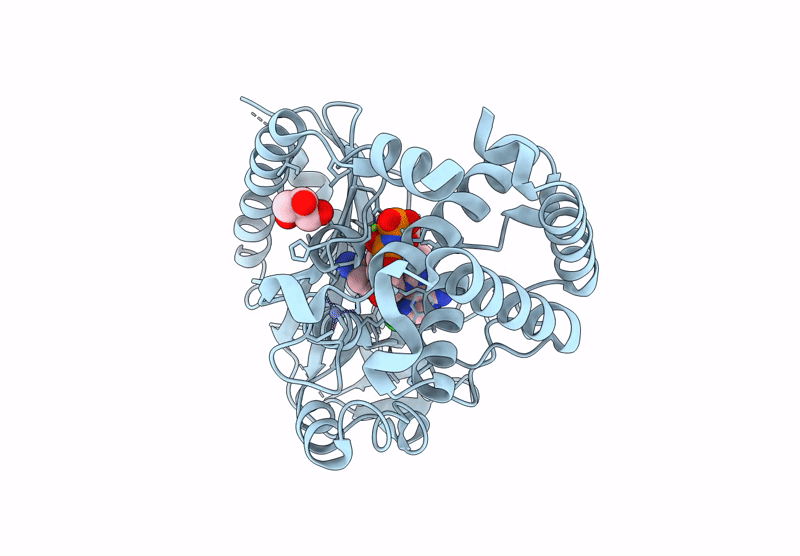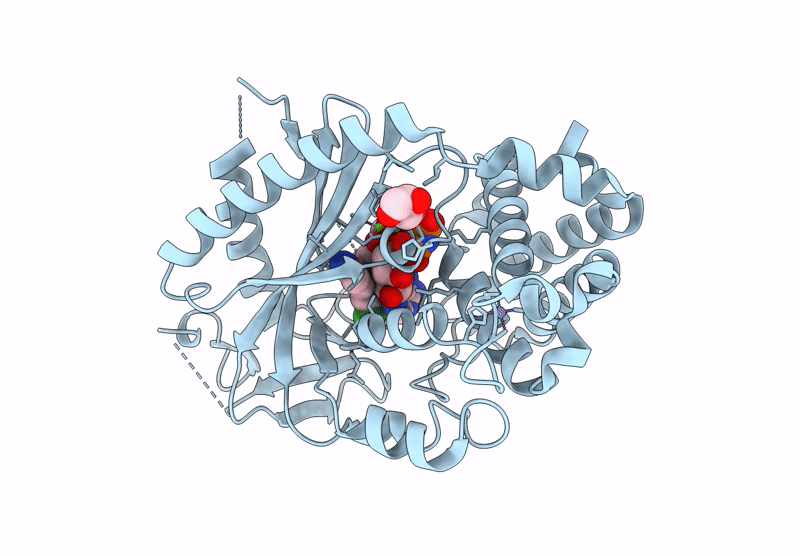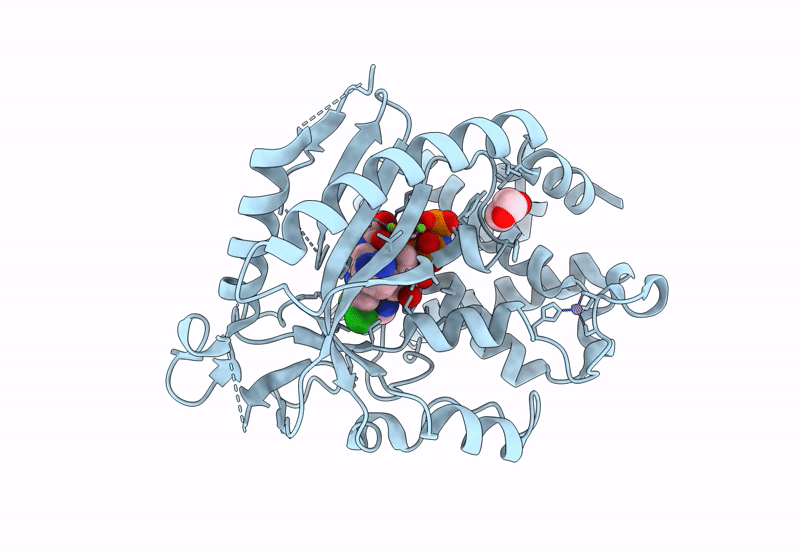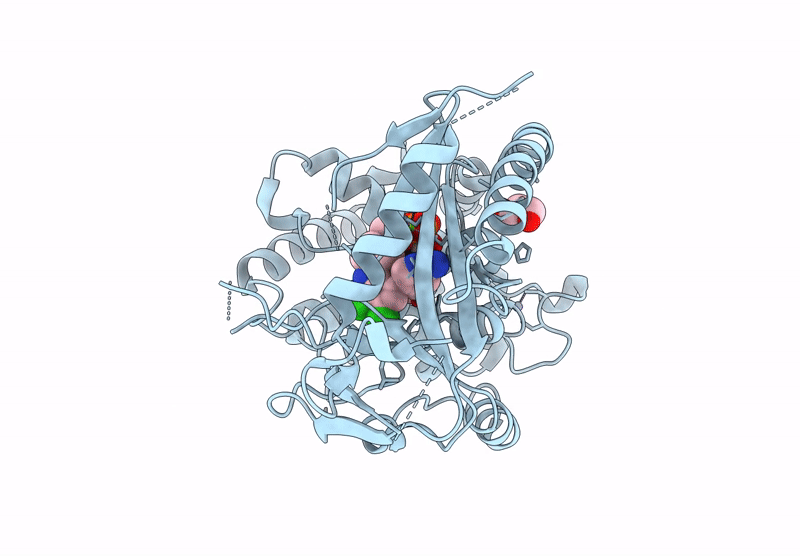
Crystal Structure Of Human Cyclic Gmp-Amp Synthase In Complex With Amppnp And Compound 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1AU3, GOL, MG, ANP, ZN |
 |
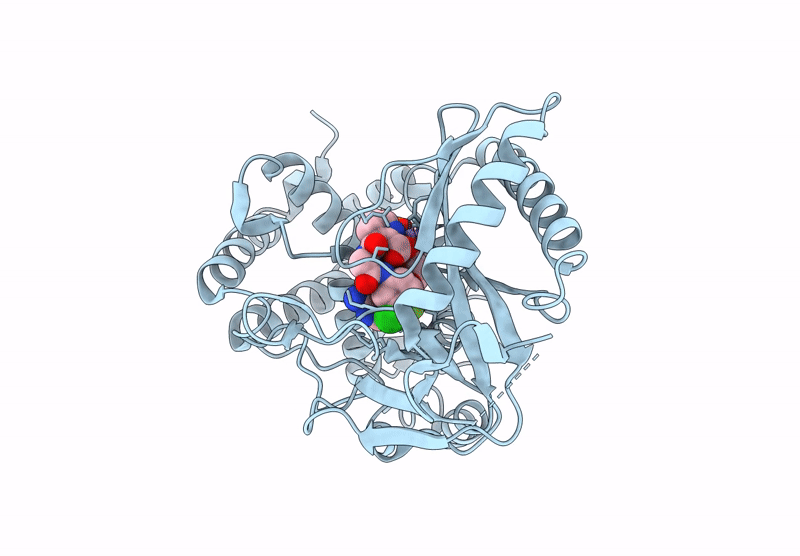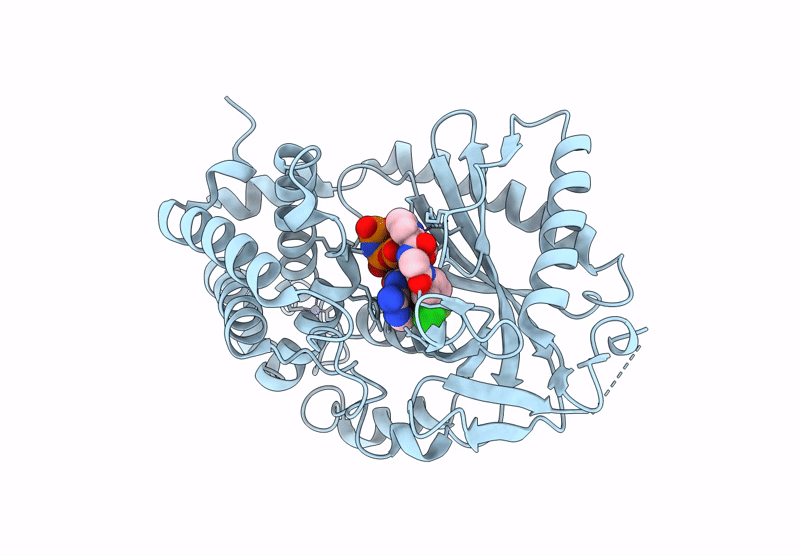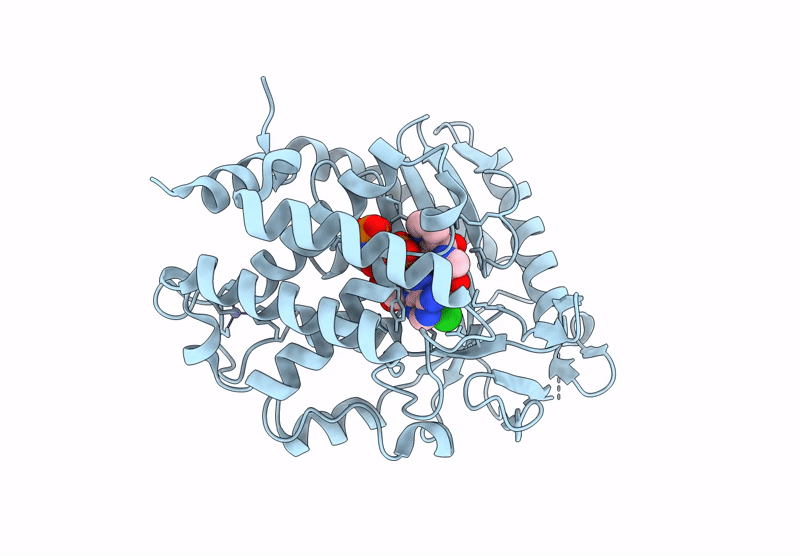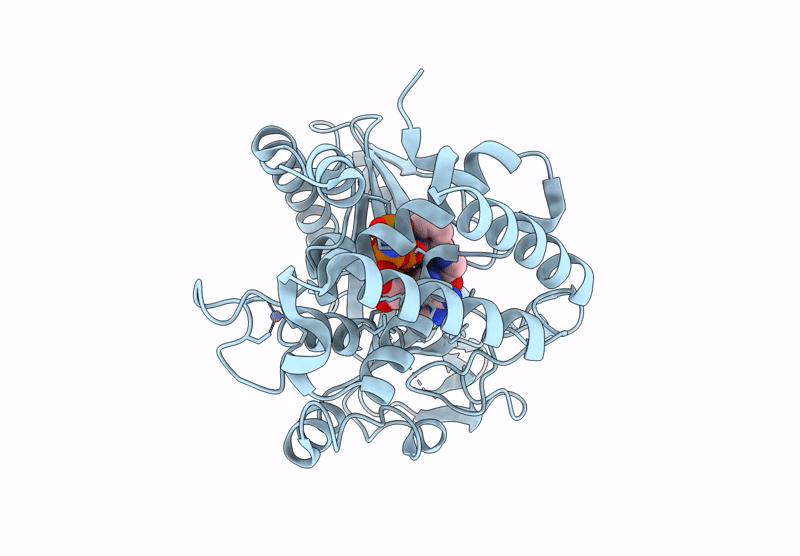
Crystal Structure Of Human Cyclic Gmp-Amp Synthase In Complex With Amppnp And Compound 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: MG, ZN, ANP, GOL, JUJ |
 |
Crystal Structure Of Human Cyclic Gmp-Amp Synthase (Cgas) In Complex With Compound 5; 3-((2-((3-Chloro-4-Fluorophenyl)Amino)-2-Oxoethyl)Carbamoyl)Picolinic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: ANP, A1BJH, ZN, MN |
 |
Crystal Structure Of Human Cyclic Gmp-Amp Synthase (Cgas) In Complex With Compound 36; (S)-(6,7-Dichloro-1-Methyl-1,3,4,5-Tetrahydro-2H-Pyrido[4,3-B]Indol-2-Yl)(5-(2-Hydroxyethoxy)Pyrimidin-2-Yl)Methanone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: ZN, A1BJA, ANP, MN |
 |
Crystal Structure Of Human Cyclic Gmp-Amp Synthase (Cgas) In Complex With Compound 23; (S)-1-(6,7-Dichloro-1-Methyl-1,3,4,5-Tetrahydro-2H-Pyrido[4,3-B]Indol-2-Yl)-2-Methoxyethan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: ZN, A1BJD, ANP, MN |
 |
Crystal Structure Of Selective Ire1A Inhibitor 29 At The Enzyme Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-05-29 Classification: TRANSFERASE/INHIBITOR Ligands: EDO, XOE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-04-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: N8S, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-04-21 Classification: TRANSFERASE/INHIBITOR Ligands: N97 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-04-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-02-27 Classification: SIGNALING PROTEIN Ligands: RPI, EDO |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-02-27 Classification: SIGNALING PROTEIN Ligands: RPI, PO4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-06 Classification: SIGNALING PROTEIN Ligands: FMT, EDO, IMD |
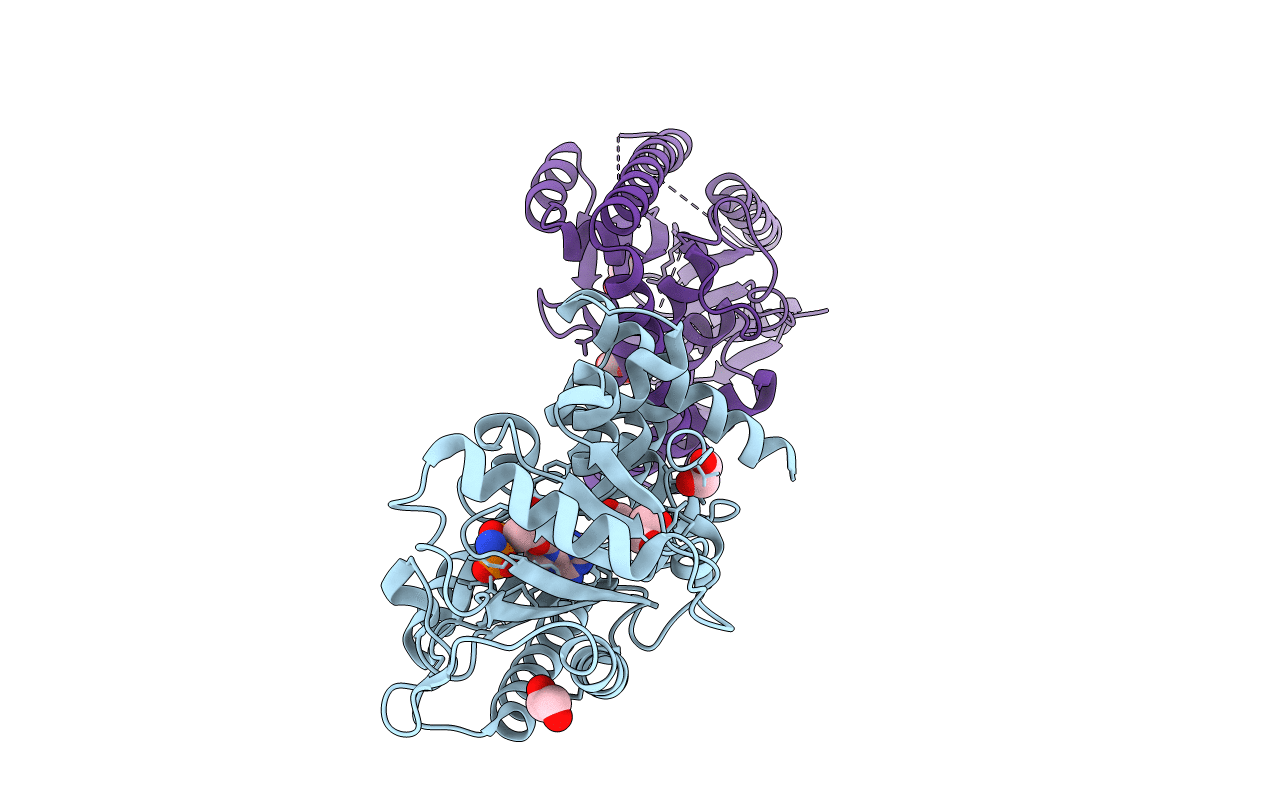 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-06 Classification: SIGNALING PROTEIN Ligands: AN2, EDO |
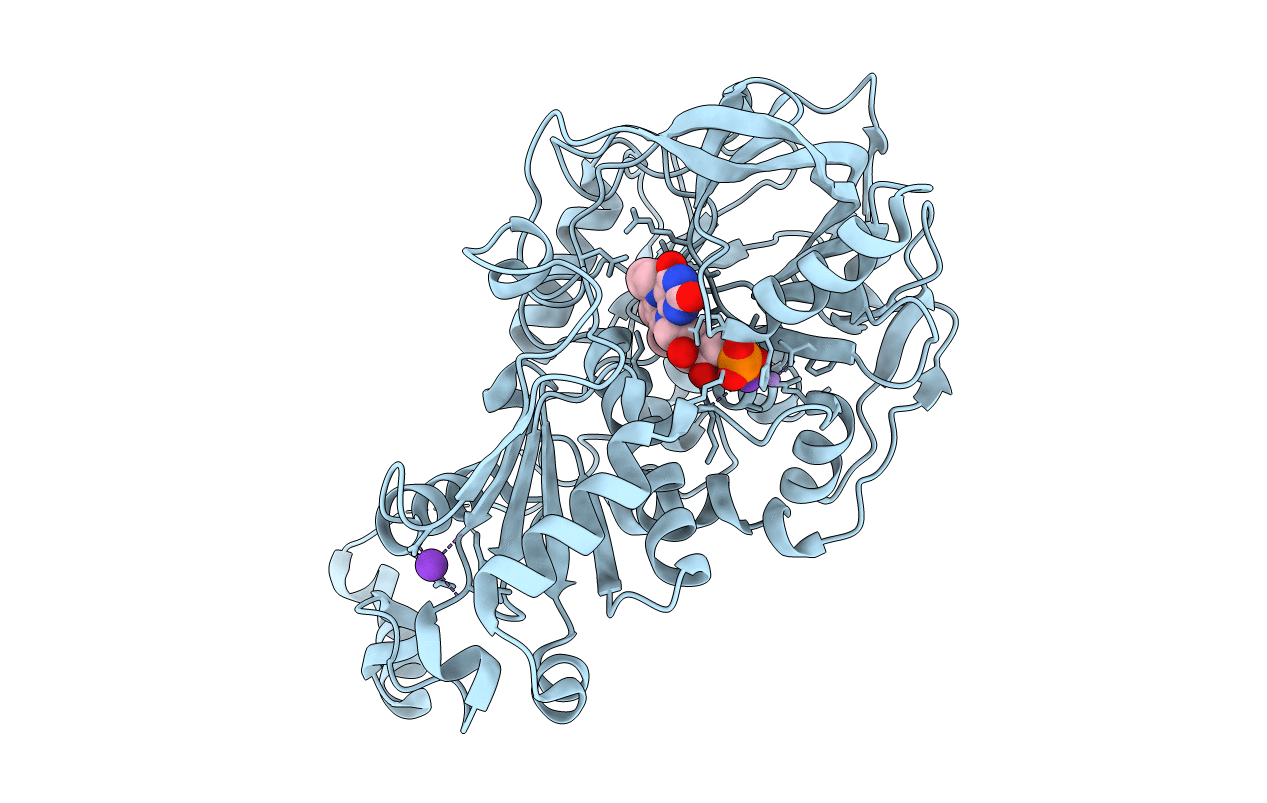 |
Structure Of A. Niger Fdc1 With The Prenylated-Flavin Cofactor In The Iminium Form.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, MN, K |
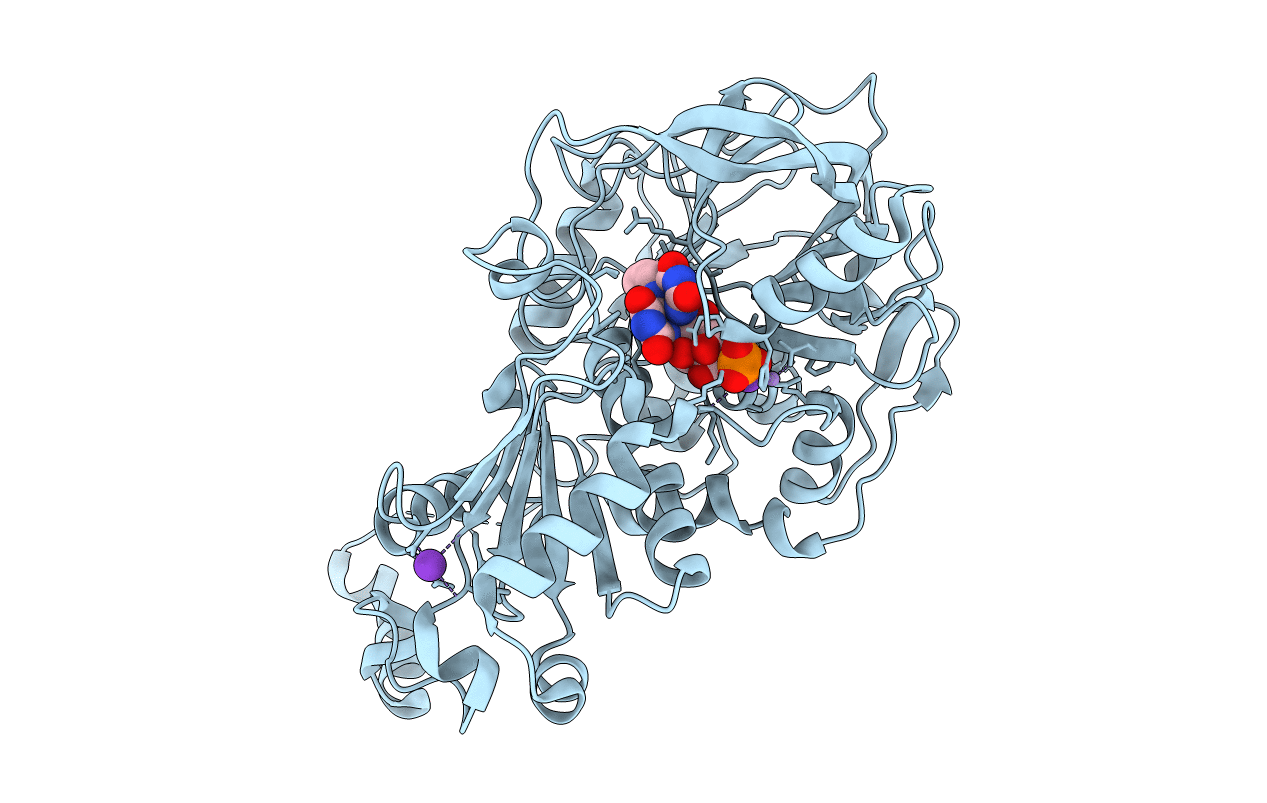 |
Structure Of A. Niger Fdc1 With The Prenylated-Flavin Cofactor In The Iminium And Ketimine Forms.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K |
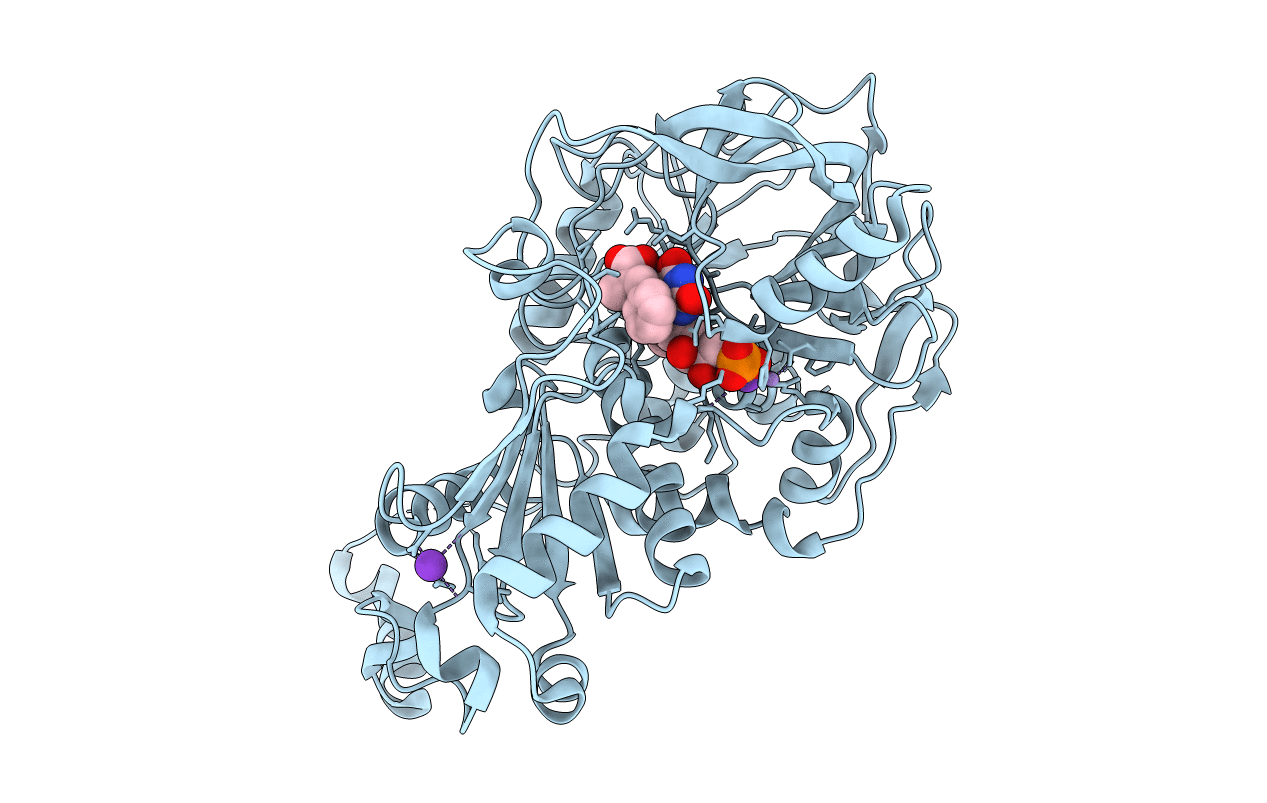 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, 4LV |
 |
Crystal Structure Of A Niger Fdc1 In Complex With Penta-Fluorocinnamic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, F5C |
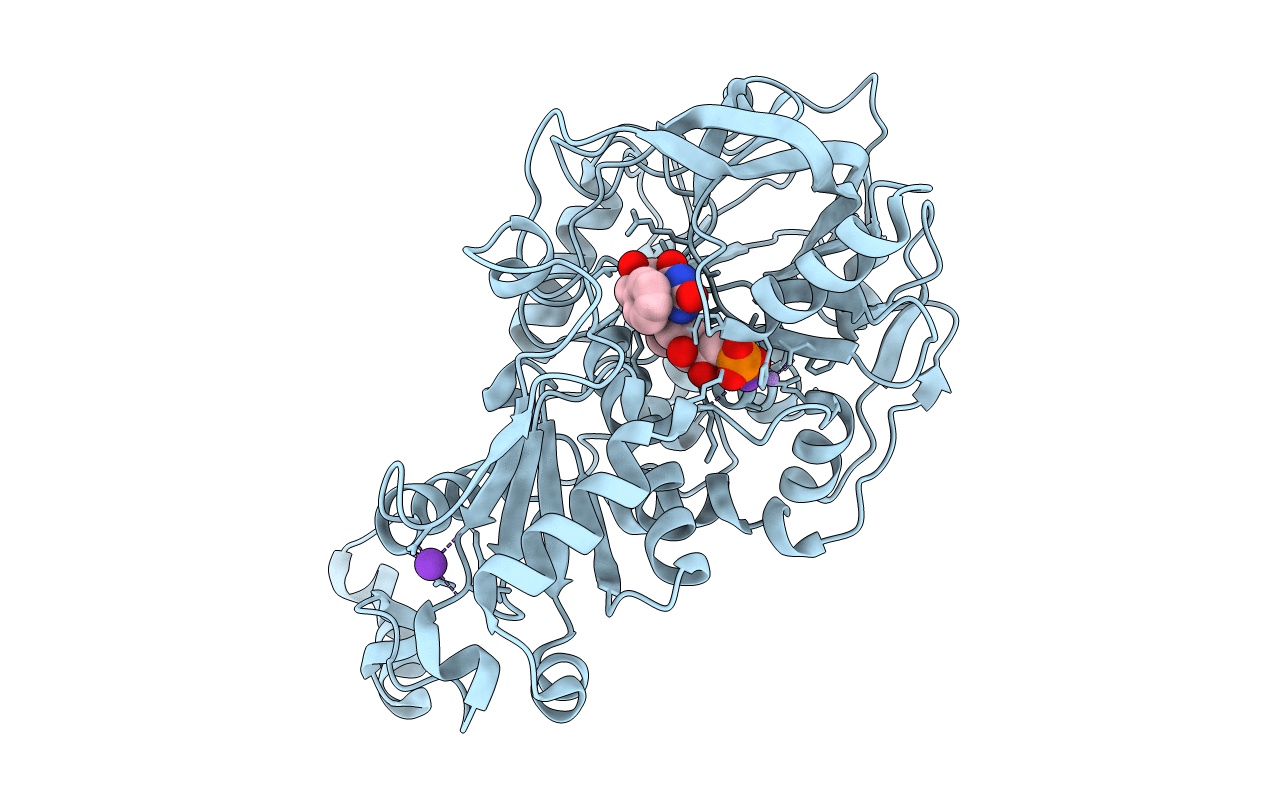 |
Structure Of A. Niger Fdc1 In Complex With A Phenylpyruvate Derived Adduct To The Prenylated Flavin Cofactor
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4MJ, MN, K |
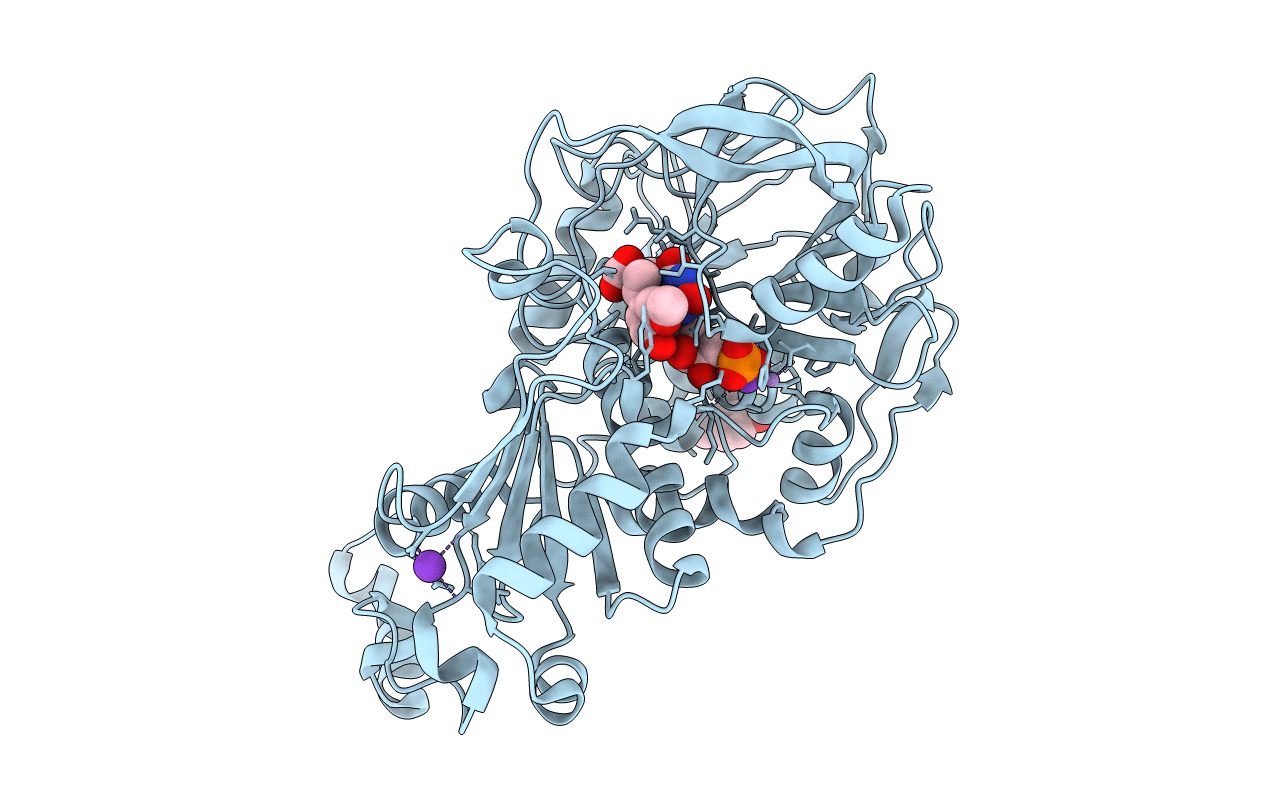 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, 4M4, CO2 |

