Search Count: 15
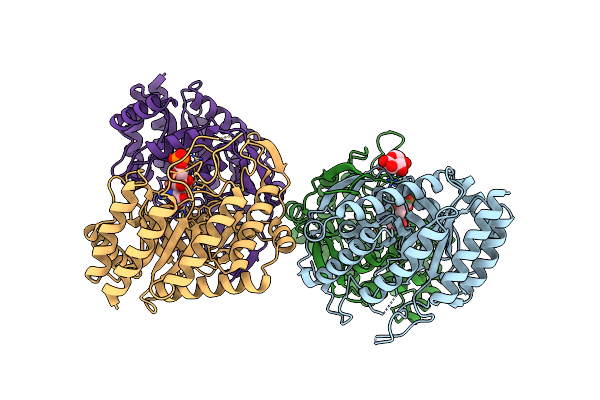 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Dahp Oxime In Unbound:(Bound)2:Unbound Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-10-12 Classification: LYASE Ligands: GLC, 52L, BGC |
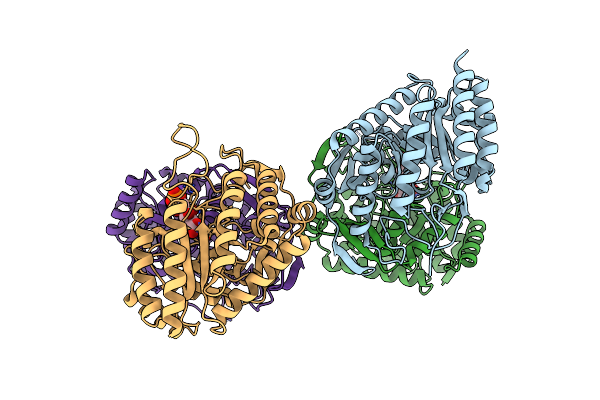 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Dahp Oxime In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-10-12 Classification: LYASE Ligands: 52L |
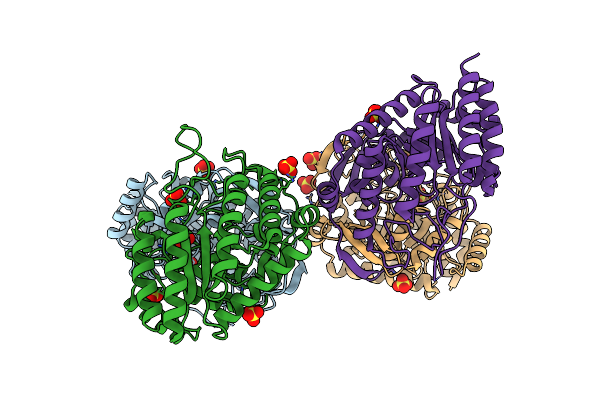 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Sulfate In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-10-12 Classification: LYASE Ligands: SO4 |
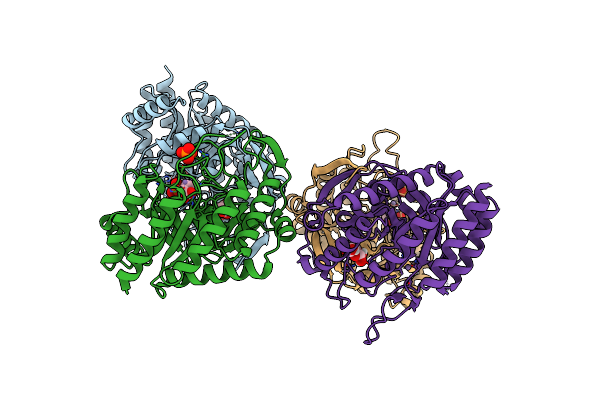 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Mn(Ii), Pep, And Pi In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-10-12 Classification: LYASE Ligands: MN, GAL, PEP, PO4 |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Mn(Ii), Pep, And Pi In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-10-12 Classification: LYASE Ligands: MN, PEP, PO4 |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Dahp Oxime, Pr(Iii), And Pi In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-10-12 Classification: LYASE Ligands: PR, FLC, PO4, ACT, 52L |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Unbound:(Bound)2:Unbound Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-11-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 7QE |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-11-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 7QH, MN |
 |
Crystal Structure Of Neub, An N-Acetylneuraminate Synthase From Neisseria Meningitidis, In Complex With Magnesium And Malate
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-10-02 Classification: BIOSYNTHETIC PROTEIN,TRANSFERASE Ligands: MG, MLT |
 |
Crystal Structure Of Metal-Free Neub, An N-Acetylneuraminate Synthase From Neisseria Meningitidis In Complex With Malate
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-10-02 Classification: BIOSYNTHETIC PROTEIN,TRANSFERASE Ligands: MLT |
 |
Crystal Structure Of Neunac Oxime Complexed With Neub, An N-Acetylneuraminate Synthase From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-02 Classification: BIOSYNTHETIC PROTEIN,TRANSFERASE Ligands: OVY |
 |
Crystal Structure Of Neub, An N-Acetylneuraminate Synthase From Neisseria Meningitidis, In Complex With Manganese, Inorganic Phosphate, And N-Acetylmannosamine (Neub.Mn2+.Pi.Mannac)
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-10-02 Classification: BIOSYNTHETIC PROTEIN,TRANSFERASE Ligands: MN9, MN, PO4 |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase In Complex With Dahp Oxime.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2016-08-10 Classification: Transferase/transferase inhibitor Ligands: GAL, 52L |
 |
Crystal Structure Of Enolpyruvyl-Udp-Glcnac Synthase (Mura):Udp-N-Acetylmuramic Acid:Phosphite From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-11-24 Classification: TRANSFERASE Ligands: PO3, EPU |

