Search Count: 64
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: 5HU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: CL, PEG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: CL, PEG, SO4 |
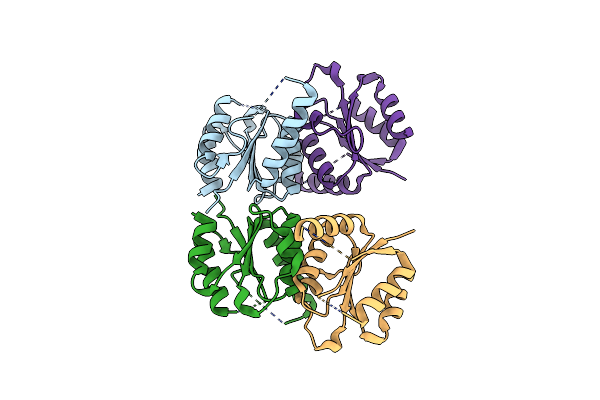 |
Structure Of Homo Sapiens 2'-Deoxynucleoside 5'-Phosphate N-Hydrolase 1 (Dnph1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-30 Classification: HYDROLASE |
 |
Structure Of Homo Sapiens 2'-Deoxynucleoside 5'-Phosphate N-Hydrolase 1 (Dnph1) Bound To Deoxyuridine 5'- Monophosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: UMP |
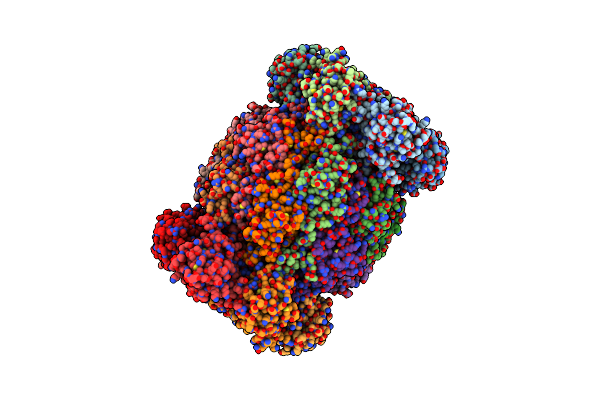 |
Leishmania Tarentolae Proteasome 20S Subunit In Complex With 1-Benzyl-N-(3-(Cyclopropylcarbamoyl)Phenyl)-6-Oxo-1,6-Dihydropyridazine-3-Carboxamide
Organism: Leishmania tarentolae
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2023-08-09 Classification: UNKNOWN FUNCTION Ligands: VYW |
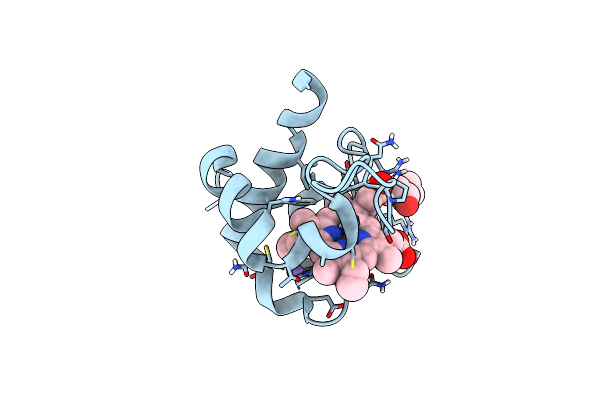 |
Organism: Cellvibrio japonicus (strain ueda107)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-07-21 Classification: ELECTRON TRANSPORT Ligands: HEC, NA, EDO |
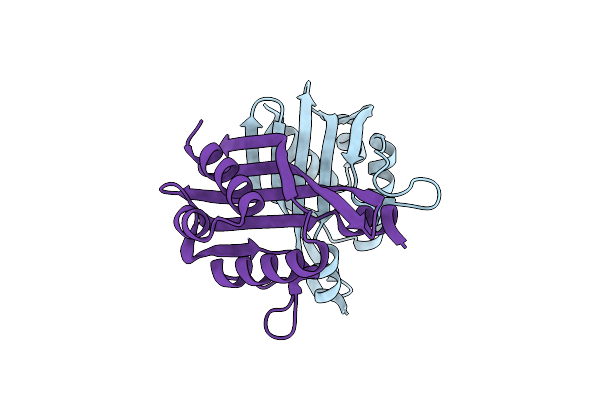 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Wild-Type Idmh, A Putative Polyketide Cyclase From Streptomyces Antibioticus
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Human Asparagine Synthetase (Asns) In Complex With 6-Diazo-5-Oxo-L-Norleucine (Don) At 1.85 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-09-18 Classification: BIOSYNTHETIC PROTEIN Ligands: ONL, EDO, EPE, CL |
 |
X-Ray Crystal Structure Of N-Acetylneuraminic Acid Lyase In Complex With Pyruvate, With The Phenylalanine At Position 190 Replaced With The Non-Canonical Amino Acid Dihydroxypropylcysteine.
Organism: Staphylococcus aureus (strain nctc 8325)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-22 Classification: LYASE Ligands: PEG |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL, SO4, NA, 54Q, DT |
 |
Organism: Staphylococcus aureus, Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, MN, GOL, NA, EVP |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, NA, GOL, MN, 53M, 53L, 50M |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-12-16 Classification: ISOMERASE |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2015-12-16 Classification: HYDROLASE Ligands: MG, GOL, MFX |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL |
 |
Crystal Structure Of The Wild-Type Staphylococcus Aureus N- Acetylneurminic Acid Lyase In Complex With Fluoropyruvate
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-11-18 Classification: LYASE |
 |
Structure And Mechanism Of Fibronectin Binding And Biofilm Formation Of Enteroaggregative Escherischia Coli Aaf Fimbriae
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-12-24 Classification: CELL ADHESION Ligands: SO4, ACT |

