Search Count: 19
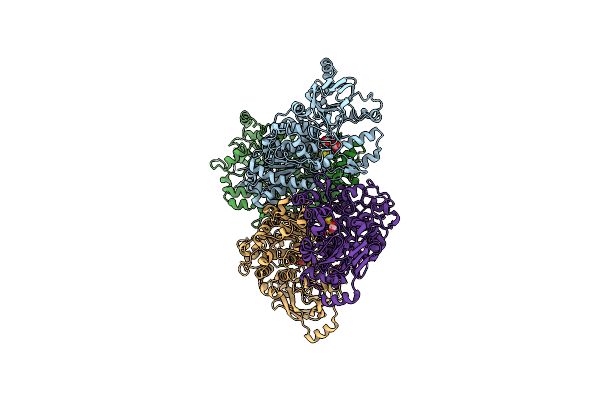 |
Crystal Structure Of Fe-S Cluster-Dependent Dehydratase From Paralcaligenes Ureilyticus In Complex With Mg
Organism: Paralcaligenes ureilyticus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-08-30 Classification: LYASE Ligands: FES, MG, BCT, CO2 |
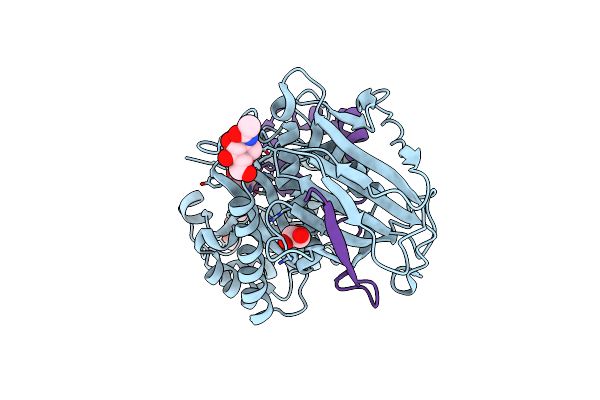 |
Crystal Structure Of Human Heparanase In Complex With Competitive Inhibitor Gd05
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: NAG, EDO, SJ5 |
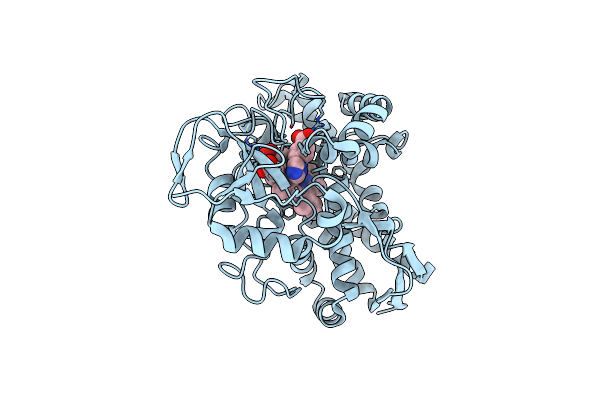 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: ZRS, HEM, CL |
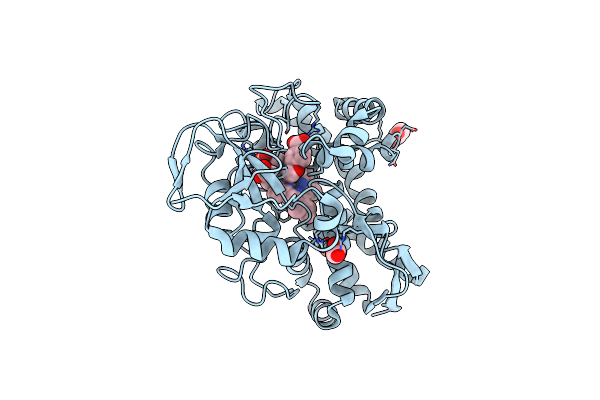 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE Ligands: ANN, CL, MG, ACT, HEM |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: CL, HEM, MW7 |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: CL, HEM, MW7 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: EPE, MGD, MO, CL, O |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: O, MGD, MO |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: HEM, PQP, CL |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: PQM, HEM, CL, ACT |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: HEM, CL, PQS |
 |
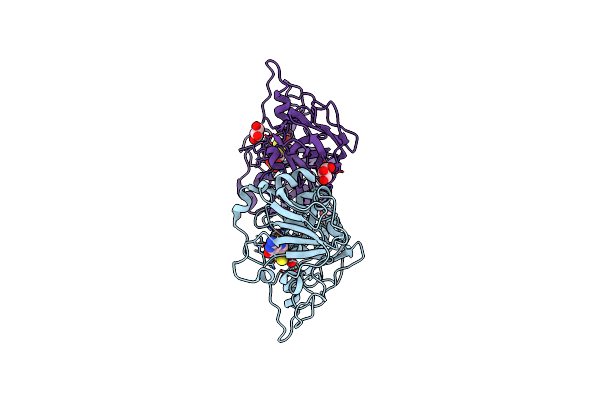
Crystal Structure Of The Sulfite Dehydrogenase, Sort R78Q Mutant From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: MSS |
 |
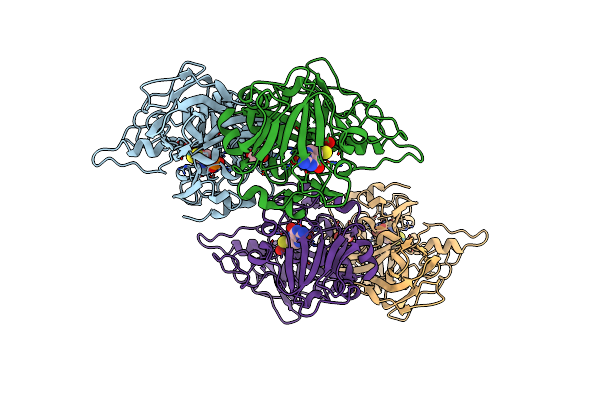
Crystal Structure Of The Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-05-24 Classification: OXIDOREDUCTASE Ligands: MSS, GOL |
 |
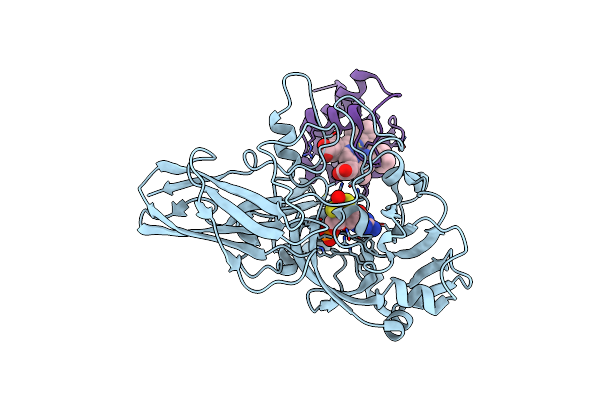
Crystal Structure Of The Sulfite Dehydrogenase Sort From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-06-03 Classification: OXIDOREDUCTASE Ligands: MSS, EDO |
 |
Crystal Structure Of The Electron-Transfer Complex Formed Between A Sulfite Dehydrogenase And A C-Type Cytochrome From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2015-06-03 Classification: OXIDOREDUCTASE/ELECTRON TRANSPORT Ligands: MSS, HEC |
 |
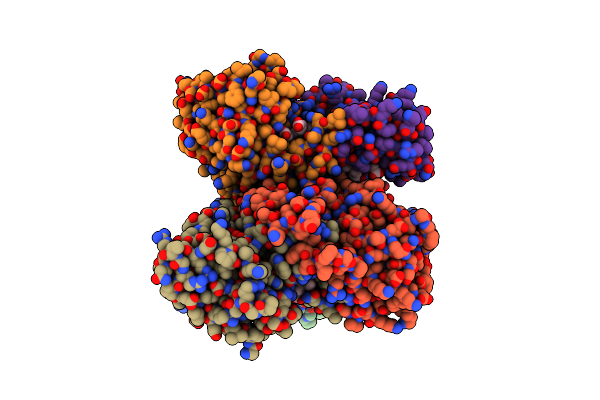
Crystal Structure Of The C-Type Cytochrome Soru From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2015-06-03 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Starkeya novella
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2011-05-18 Classification: Heme-binding protein/Heme-binding protein Ligands: HEC, SO4 |
 |
Organism: Starkeya novella
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-05-18 Classification: Heme-binding protein Ligands: HEC |
 |
Structure Of Bacterial Globin From Campylobacter Jejuni At 1.35 A Resolution
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2010-02-16 Classification: OXYGEN TRANSPORT Ligands: HEM, CYN |

